- Submissions

Full Text
Modern Concepts & Developments in Agronomy
Assessing Wheat Damage Caused by Gaeumannomyces graminis Using Plant Pathology Techniques, Image Analysis, and Satellite Data Image Analysis
I Vagelas*
Assistant Professor, Department of Agriculture Crop Production and Rural Environment, University of Thessaly, Greece
*Corresponding author:I Vagelas, Assistant Professor, Department of Agriculture Crop Production and Rural Environment, University of Thessaly, Greece
Submission: March 31, 2021Published: May 06, 2021

ISSN 2637-7659Volume8 Issue 4
Abstract
This paper discusses the potential use of image analysis and satellite images to provide results at a plant and a wheat-field level of damage caused by Gaeumannomyces graminis, the Take-all fungus of wheat. Type of G. graminis specimen and wheat-field damage were examined and described in this research with plant pathology techniques. Obtained results of those techniques were further analyzed with image acquisition and satellite images derived at a plant and a wheat-field respectively. With those images, we provided counts and data for assessing wheat Take-all disease, at a plant and wheat-field level.
Keywords: Take-all; Crop canopy damage; Acquisition; Precision agriculture
Introduction
Gaeumannomyces graminis the Take-all fungus is the most specific disease problem of wheat worldwide. The fungus colonized at the beginning of the season the wheat plant roots (primary infection) associated with chlorotic patches of plants (initial symptoms). Infected prematurely plants eventually killed in these patches and appear stunted off-color causing whiteheads. These obvious late-season symptoms (whiteheads) appeared within circular patches in the infected wheat field and were recognized by growers. The Take-all fungus is soil-borne, survives saprophytically in crop residue is particularly prevalent in second wheat crops [1], attacks the root system of the wheat emerging seedlings, and causes Takeall diseases. However, the infection caused by the fungus can increase under high moisture during the autumn season (personal observations).
Based on the chlorotic patches of wheat infected plants observed early in the season associated with symptoms of root rot [2], G. graminis early infection can be easily recognized. At first, the symptomatic roots are short and rotted and stolons are lifted with black lesions above the ground. Gradually, the entire root turns black, and the fungus is transmitted and spread to the nearest plants which forming new roots as a root-to-root infection allow the appearance/epidemics of the disease associated with Take-all patches. In these patches when the period of crop maturity is under wet conditions (late April, personal observations), in most cases, fungus produced perithecia at the base of infected culms. As literature shows [3,4], the ascospores length of those perithecia are important in distinguishing varieties (var.) of G. graminis associated with the Take-all disease as var. tritici or avenae usually associated with Take-all of wheat from var graminis which is not [3]. That point is one objective of this paper to reports ascospore length measurement of G. graminis isolates from central Greece.
Moreover, as the disease on the root surface and stem base appear as brown to dark discoloration these disease margins can easily estimate and measure using image analysis technology, the other objective of this study.
Finally, if the fungus is present in the field for many years it is recommended to use that it is possible to detect and to analyze the patches of whiteheads produced after the anthesis period [1]. We assume that those patches can easily detect using satellite images, the third objective of this study.
Material and Methods
Gaeumannomyces graminis isolates
In late April of 2015 patches of wheat heads, were seen in winter wheat crops grown in central Greece. From those patches, plants were collected, washed free of soil, and isolates were obtained from surface-sterilized tissues plated on potato dextrose agar (PDA) to identify the caused agent of wheat damage. As our data showed that the wheat damage caused by the fungus G. graminis, types of hyphopodia, and ascospore size [5], were used as specific characteristics identification in recognizing varieties of G. graminis [3]. Perithecia (n=230) were collected from the base of infected plants as described by [3] and measure with a microscope.
Sampling and disease assessment
In late April of 2016 eight sampling fields in central Greece, showing the previous year’s severe Take-all disease were chosen. Those fields were marked with a GPS device and samples were taken from patches of wheat heads. Samples were collected randomly in each field, from 20 patches of wheat heads, each patch marks as a 1x1m subplot, and six plants were sampled from each of the 20 subplots/sampling field [6]. Sampled plants were washed free of soil and examined for Take-all by isolated and identified (microscopic), the fungus from diseased tissues of the plants. The disease was assessed on root symptoms by a root rot index 0 to 5, where 0=no disease and 5=severe root rot (heavy infection of roots and stem bases).
Further, disease incidence (D.I.) was evaluated using the following calculations, D.I. (%) = [(Number of Infected Plants/Total Number of Plants) x 100. Classification method of infection were graded as, slight infection (SI), D.I. <40; moderate infection (MI), 40 < D.I. < 65; severe infection (SevI), D.I. > 65 (Table 1).
Table 1: Estimate of Take-all root rot index, and disease incidence (D.I.), in eight (A1-A8) wheat fields.
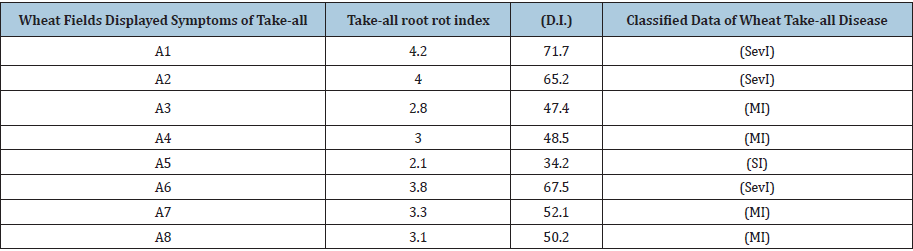
Image analysis
Heavily infected plants with Take-all root rot diseases, appeared dark brown discoloration symptoms at the roots and the basal stem was used in this study. Images were taken from those plants (20 patches described above) and compared with images taken from unaffected (control) plants. The images were processed in the “ImageJ” image analysis program. The discoloration, the Disease Symptoms (D.S.), was used as a parameter to estimate in percent the disease caused by the pathogen. That color contrast in the images was enhanced by applying the “Enhance Contrast” tool. We cropped the plant from the background by using “selection” and “clearoutside” tools.
The cropped image was visualized in RGB space and threshold values for green pixels left in the image were determined by “colour threshold tool”. The “threshold colour macro” written by Landini et al. [7] was applied to select the range of red, green, and blue components of the color such that green pixels of background were separated from plant pixels. To identify white pixels of fungus, affect the image was viewed in HSB space. The range of hue, saturation, and brightness values for white pixels were determined by applying the same macro. The threshold colour macro was modified so that it takes the above-mentioned range of values as input, separates particular colour in the image, and counts the number of pixels of the specific colour. The pixels count was converted to percentage effect in green color. The percent discoloration (infected plants) was estimated from the pixels of the whole plant, background green color, an unaffected part of the plant.
Satellite imagery. normalized difference vegetation index
We used the normalized difference vegetation index (NDVI) derived from satellite images from the above-mentioned eight wheat fields (Figure 1), as described by Yengoh et al. [8]. Two wheat fields (C1, C2, Figure 1) with no Take-all symptoms were selected as the experimental control wheat fields. In our experiment (Figure 1), NDVI values near to zero mean no vegetation, close to 0.4 (0.3- 0.5) indicates significant disease infection, and close to +1 (0.7-0.9) indicates the highest possible density of green wheat leaves.
Figure 1:Spectral (NDVI) differences between, the healthy wheat fields (C1, C2) and wheat fields with visible symptoms of the Take-all disease (A1-8).
Result and Discussion
Gaeumannomyces graminis isolates
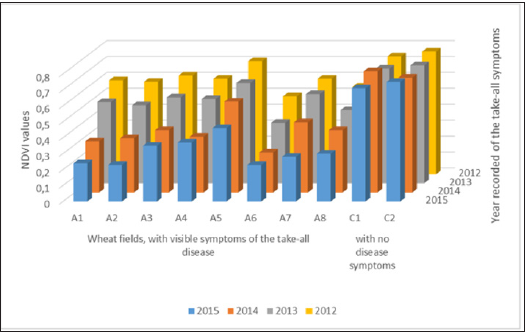
The morphological characteristics (Figure 2) of G. graminis fungus isolates, obtained from diseased wheat fields from central Greece, had Perithecia length 280-460μm, ascus length 85-132μm, ascospore length 75-105μm and did not form lobed hyplodia and were, therefore, G. graminis var. tritici as described by Walker [9].
Figure 2: Morphological features of Gaeumannomyces graminis var. tritici:
A. Perithecium.
B. Asci and ascospores.
C. Simple hyphopodia.

Sampling and disease assessment
Our results (Table 1) showed that the pathogen produces moderate to severe infection to wheat plants indicate that the pathogen occurs for several years. This infection is significant differences between wheat fields (Table 1). Heavy infection of roots and stem bases was observed in wheat fields with high Take-all Disease Incidence (D.I.) values (Table 1).
Image analysis
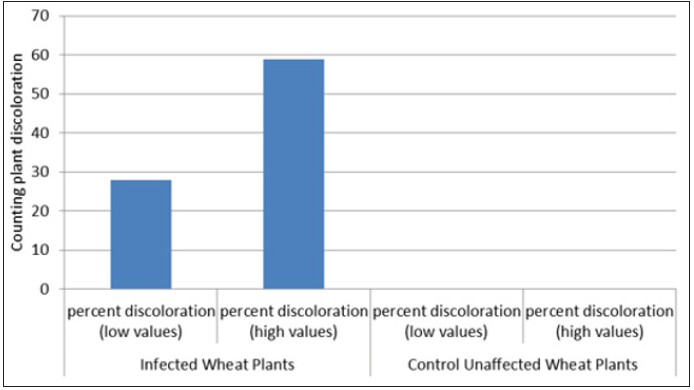
The image analysis (Figure 3), indicate that the percent of discoloration values on infected wheat plants (blocks) with G. graminis fungus, ranges from 28 to 59 percent compared with zero percent appeared in control blocks (Figure 4). Those discoloration results (Figure 3 & 4), show very close values to those presented in Table 1. Overall, image analysis (Figure 3), exhibits as an appropriate tool to explain the disease incidence values of the wheat (Table 1), caused by G. graminis fungus.
Figure 3:Various masks obtained in procedure of counting discoloration plant effect.
A. Binary mask of the whole plant.
B. Background green pixels.
C. Binary mask of the unaffected part of the plant.
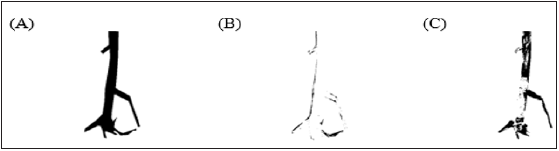
Figure 4:Image analysis. Percent of discoloration values on infected, and unaffected wheat plants with Gaeumannomyces graminis fungus.
Satellite imagery. normalized difference vegetation index
Figure 1 shows that, from all the ten wheat fields, the lowest NDVI values were obtained from the eight wheat fields with the visual Take-all disease symptoms. The spectral (NDVI) differences for years 2015, 14, 13, and 12 between, the wheat fields with visible symptoms of the Take-all disease and healthy wheat fields (C1, C2) were significantly lower, provided evidence that the fungus appeared for many years in the eight observed wheat fields. Data shows that the healthy wheat fields C1, C2 (Figure 1), absorb most of the visible light that hits them (Figure 1). Overall, we concluded that this method is based on the optical properties of wheat plants canopy and can be used for identifying plants diseases stress [10] or fungus damage such as G. graminis.
Conclusion
As the results showed, Gaeumannomyces graminis var. tritici is the prenominal pathogen that caused the Take-all wheat disease in central Greece. Finally, we conclude that plant image analysis and satellite imagery can use as a useful tool for
i. disease classification,
ii. disease detection, and
iii. disease outbreaks by year.
References
- Macdonald AJ, Gutteridge RJ (2011) Effects of take-all (Gaeumannomyces graminis tritici) on crop N uptake and residual mineral N in soil at harvest of winter wheat. Plant and Soil 350(1-2): 253-260.
- Elliott ML (1993) Association of Gaeumannomyces graminis graminis with a St. Augustinegrass root rot disease. Plant Disease 77(2): 206-209.
- Yeates JS (1986) Ascospore length of Australian isolates of Gaeumannomyces graminis. Transactions of the British Mycological Society 86(1): 131-136.
- Freeman J, Ward E (2004) Gaeumannomyces graminis, the take-all fungus and its relatives. Mol Plant Pathol 5(4): 235-252.
- Walker J (1972) Type studies on Gaeumannomyces graminis and related fungi. Transactions of the British Mycological Society 58(3): 427-457.
- Gosme M, Lucas P (2010) Effect of host and inoculum patterns on take-all disease of wheat incidence, severity and disease gradient. European Journal of Plant Pathology 129(1): 119-131.
- Landini G, Randell DA, Fouad S, Galton A (2017) Automatic thresholding from the gradients of region boundaries. J Microsc 265(2): 185-195.
- Yengoh GT, Dent D, Olsson L, Tengberg AE, Tucker CJ (2015) Challenges to the use of NDVI in land degradation assessments. Springer Briefs in Environmental Science, pp: 55-56.
- Walker J (1981) Taxonomy of take-all fungi and related genera and species. In: Biology and control of take-all. Asher MJC, Shipton PJ (Eds.), Academic Press, New York, USA, pp: 15-74.
- Pourazar H, Samadzadegan F, Dadrass Javan F (2019) Aerial multispectral imagery for plant disease detection: Radiometric calibration necessity assessment. European Journal of Remote Sensing 55(3): 17-31.
© 2021 I Vagelas. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






