- Submissions

Full Text
Modern Concepts & Developments in Agronomy
Aggressiveness and Genotyping of Phytophthora Infestans Isolates from Nicaragua
Blandón Díaz JU1,2*, Högberg N2, Grönberg L2, Widmark AK2 and Yuen J2
1Department of Plant and Forest Protection, National University of Agriculture (UNA), Managua, Nicaragua
2Swedish University of Agricultural Sciences, Dept of Forest Mycology and Pathology, Sweden
*Corresponding author: Blandón Díaz JU, Department of Plant and Forest Protection, National University of Agriculture (UNA), Managua, Nicaragua
Submission: February 23, 2021Published: March 10, 2021

ISSN 2637-7659Volume8 Issue 2
Abstract
The late blight pathogen, P. infestans, is a hemi biotrophic, heterothallic, diploid oomycete that remains a major constraint for potato and tomato production worldwide and causes economic losses yearly calculated to be over four billion euros. This study was undertaken to test the following hypotheses: i) P. infestans isolates from Nicaragua do not differ in their aggressiveness toward potato or tomato; and ii) the P. infestans population from Nicaragua is formed exclusively by a single clonal lineage. Five SSR multilocus genotypes among 72 isolates of P. infestans were detected and all of them were of the Ia mtDNA haplotype and A2 mating type. The NI-1 single multilocus genotype continued to be dominant within the clonal lineage. The genotypic analysis revealed neither host specificity nor population differentiation between potato and tomato isolates. Aggressiveness tests revealed that tomato isolates showed a general, but not exclusive, host-specificity and were more aggressive on tomato. In contrast, potato isolates showed host-preference toward tomato leaflets and were more aggressive on them. The findings of this study provide useful information regarding late blight disease management on potato and tomato crops in Nicaragua.
Keywords: Quantitative pathogenicity; Plant pathogen diversity; Molecular markers; Population structure
Introduction
The late blight pathogen, P. infestans, is a hemi biotrophic, heterothallic, diploid oomycete that remains a major constraint for potato and tomato production worldwide and causes economic losses yearly calculated to be over four billion euros [1]. In compatible interactions with potato, the biotrophic phase of P. infestans can last from three to five days, after which macroscopic symptoms are evident (necrotrophic phase). In tomato leaves, an extended period of biotrophy has been observed due to a compatible interaction with tomato-specialized isolates [2-4]. As a heterothallic oomycete, P. infestans possesses two compatibility groups, referred to as mating types A1 and A2, which enables the production of oospores in locations where both A1 and A2 mating types are present contributing in that way to its overwintering survival and shaping the nature of its population structure [5]. In locations where only the asexual cycle is found, P. infestans survives as mycelium in infected potato tubers and debris [6] and also probably in alternate wild hosts.
Prior to the 1980s, worldwide populations of P. infestans were dominated by a single clonal lineage known as the US-1 “old” genotype, having the A1 mating type [7,8]. On the contrary, in the Toluca Valley in central Mexico the A1 and A2 mating types were present in approximately equal frequencies and the populations of P. infestans were entirely different from populations in other locations [5,9]. Since the mid-1980s changes in the population structure of P. infestans outside Mexico have been reported [10]. These changes brought about the displacement of the ‘old’ genotypes by ‘new’ ones, which are characterized by increased fitness and aggressiveness in addition to metalaxyl resistance [11]. Migration has played an important role in the observed changes in global population structure of P. infestans in the latter part of the last century [5]. In some areas where both mating types are present, it has been shown that late blight epidemics start earlier than before due to the presence of oospores as initial inoculum. There are also reports of more aggressive isolates, which has resulted in an increase in the use of fungicides for the disease control [12,13].
Suggested management strategies for this disease include use of clean seed, elimination of real and potential sources of inoculum (infected cull piles, volunteer potato plants, and wild alternate hosts), fungicides, Decision Support Systems (DSS), intercropping, cultivar mixtures and extended crop rotations (3-4 years) to avoid early infections developed from oospores. Along with these strategies, the use of resistant cultivars against late blight is of utmost importance, especially in locations where environmental conditions are conducive for disease development and potato growers cannot afford the numerous fungicides required to control the disease [5,12,14-16]. However, the development and use of resistant cultivars may depend on the variability of the pathogen, including isolate aggressiveness. In plant-pathogen interactions, “aggressiveness” or quantitative pathogenicity [17], refers to the ability of a given pathogen to induce severe epidemics on compatible hosts. The quantitative traits frequently used to measure aggressiveness are infection efficiency, latency period, sporulation rate, infectious period and lesion size [18]. Differential interactions for aggressiveness components between current population of P. infestans and cultivars have been studied [15,19-21].
The host range of P. infestans, generally, has been considered to be restricted to two important crops, potato and tomato, and various solanaceous weeds [22], but the factors determining this host range remain unknown [23]. P. infestans has been reported affecting various species in the genus Solanum and also non solanaceous host plants [24-26]. Some degree of pathogenic specialization of P. infestans to potato or tomato has been shown in the early 1900’s and later [2,4,5,27-29]. In USA, for example, the US-8 genotype has been detected to occur on potatoes and the US-7 and US-17 genotypes have been recovered from tomatoes [30]. A study in Kenya and Uganda clearly showed that late blight epidemics in potato and tomato were caused by two separate, host-adapted populations of P. infestans [4]. On the contrary, in a recent study carried out in Taiwan, no host specificity on potato or tomato among P. infestans isolates from tomato was found [31]. Negative relationships (genetic trade-offs) between qualitative and quantitative traits required to infect one or another host can drive the appearance of pathogenic specialization mediated by antagonistic pleiotropy, in which one or more genes favour pahtogen’s performance in one host, but impair its performance in another [18,32,33].
In a previous study conducted with P. infestans isolates from Nicaragua, no evidence of population differentiation between isolates collected from potato and tomato plants was found, based on phenotypic (mating type, virulence and fungicide sensitivity) and genotypic analysis [Simple sequence repeats markers (SSRs, also known as microsatellites) and mitochondrial DNA (mtDNA) haplotyping]. Consequently, it was concluded that Nicaraguan population of P. infestans belonged to a single clonal lineage which was dominated by the NI-1 genotype. However, in the previous work aggressiveness tests were not conducted, hence, it is not known if there is any difference in aggressiveness between potato and tomato isolates in Nicaragua. Therefore, this study was undertaken to test the following hypotheses: i) P. infestans isolates from Nicaragua do not differ in their aggressiveness toward potato or tomato; and ii) P. infestans population from Nicaragua is formed exclusively by a single clonal lineage.
Material and Methods
Phytophthora infestans cultures
Potato and tomato leaflets with a single lesion of P. infestans were collected in Nicaragua from July 2009 to January 2010. Tomato fruits with initial symptoms of late blight were collected as well. Samples were taken from commercial fields in the main potato and tomato growing areas of Nicaragua. Infected leaflets with a single lesion were washed with distilled water and dried with filter paper. Thereafter, they were individually placed upper surface down in inverted Petri dishes lined with 1.5% water agar in the base and incubated at 18 °C to promote sporulation. Mycelia and sporangia were then transferred to a pea agar medium [34], amended with antibiotics (0.2g ampicillin and 10mg pimaricin L-1) and incubated at 18 °C in darkness. One week later, the isolates were transferred to a pea agar medium without antibiotics and incubated at 18 °C for growth and sporulation. Seventy-two isolates (53 from potato and 19 from tomato leaflets and fruits) of P. infestans were obtained. These isolates were kept at 12 °C and transferred to rye pea agar [(30g organic rye, 60g frozen green peas, 10g sugar or sucrose, 15g agar and 1L deionised water); [35]] prior to their use in the different tests. All of the 72 isolates were used for SSR analysis, mtDNA haplotyping and mating type determination. A subset of 31 isolates (16 potato isolates and 15 tomato isolates) was used for aggressiveness testing.
Mating type determination
Mating type was determined by pairing each unknown isolate individually with known tester isolates of A1 and A2 mating type on rye pea agar. Hyphal interaction zones were microscopically observed after 7-14 days incubation at 18 °C in darkness. Oospores were produced in the margins of opposite mating types. Isolates that produced oospores with the known A1 tester isolates were designated as the A2 mating type and isolates that produced oospores with the known A2 tester isolates were designated as the A1 mating type.
Plant material and inoculum preparation
Potato plants (cv. Bintje) and tomato plants (cv. Homestead 24) were grown in 3L plastic pots filled with a soil mixture (pea-sand-clay-lime-compost), in a greenhouse at 18-20 °C (night/day temperature) and 16 h photoperiod. Plants were watered weekly with a fertilizer solution [N(8%)-P(3%)-K(5%)] supplemented with micronutrients. These plants were used to test the pathogenicity of the potato and tomato isolates on their host of origin as well as to produce inocula for the subsequent tests. Detached leaflets of 6-8 weeks old potato plants were individually placed, abaxial surface up, on the lids of inverted Petri dishes lined with 1.5% water agar acting as humid chambers. Sporangia from 2-week-old potato isolates in Petri dishes were collected in distilled water with a paintbrush. Prior to inoculation, sporangial suspensions were kept at 4 °C for 2h to release zoospores. Thereafter, each potato leaflet was inoculated with the mixture of sporangia and zoospores. Each leaflet was covered as evenly as possible with 20µL droplets of the sporangia/zoospore suspension to ensure infection. Inoculated potato leaflets were incubated at 16 °C (day/night temperature) and 90% relative humidity. After a week of incubation, sporangia were collected from the infected leaflets with a paintbrush in distilled water and sporangial concentration was adjusted to 2x104 sporangia mL-1 using a hemocytometer. The resulting sporangial suspensions were further used to inoculate potato leaflets and determine the aggressiveness parameters. The same procedure was followed to test the pathogenicity of tomato isolates in tomato leaflets and to obtain the appropriate (pathogenic) inoculum with the abovementioned sporangial concentration.
Aggressiveness determination
Sixteen isolates from potato and fifteen isolates from tomato were used in these cross-inoculation assays, i.e., potato leaflets were individually inoculated with potato and tomato isolates and the same was done with tomato leaflets. The inoculum was prepared directly from infected potato and tomato leaflets as described above. Each isolate-host combination was repeated five times (one leaflet of potato or tomato per Petri dish). Potato or tomato leaflets were placed abaxial side up on the lids of inverted Petri dishes lined with 1.5% water agar in the base and inoculated with a 20µL droplet of sporangial suspension adjusted to 2x104 sporangia mL-1 of the appropriate test isolate (potato or tomato). Thereafter, the inoculated leaflets were incubated at 16 °C and 16h day length. The incubation period (IP), latency period (LP), lesion area (LA), lesion growth rate (LGR), spore production (SP), sporulating area (SA) and sporulation rate (SR) were determined as described elsewhere [28,36,37]. An aggressiveness index (Ai) for each isolate-host combination was calculated using the following equation: Ai=ln (LA x SP x 1/LP) [21,38].
SSR fingerprinting and mtDNA haplotyping
Microsatellite analysis was carried out using seven loci: Pi4B, PiG11 [39] and Pi16, Pi70, PiD13, Pi63 and Pi04 [40]. Forward primers 4B, Pi16, D13 and Pi04 were labelled with 6-FAM (TAG Copenhagen), whereas G11, Pi70 and Pi63 were labelled with NED (Applied Biosystems). PCR amplifications were performed in 15µL containing approximately 10ng of genomic DNA, 0.2mM dNTPs, 0.4µM of each forward and reverse primers, 0.04U µL-1 ThermoRed DNA polymerase (Saveen & Werner AB) and 1X reaction buffer Y (containing 2mM MgCl2) supplied by the manufacturer. For the primers for locus 4B and Pi70, 4mM MgCl2 was used. The PCR conditions were as follows: an initial denaturation at 94 °C for 3min, followed by 30 or 33 cycles (dependent on the primers) at 94 °C for 30s, 30s annealing temperature ranging from 50 °C to 62 °C dependent on the primers, elongation at 72 °C for 1min and final extension at 72 °C for 25min (GeneAmp® PCR System 2700, Applied Biosystems). The annealing temperature and the number of cycles for each primer were as follows: 50 °C and 33 cycles for primer D13, 58 °C and 33 cycles for primers Pi4B and Pi70, 60 °C and 33 cycles for Pi63, 60 °C and 30 cycles for Pi63, 60 °C and 33 cycles for Pi16 and 62 °C and 30 cycles for primers PiG11 and Pi04. For dried leaflet samples, the annealing temperature and the number of cycles for primer Pi63 were 58 °C and 33 cycles respectively. Separation of the amplified fragments was done using an ABI 3730xl DNA analyzer at Uppsala Genome Center, Rudbeck Laboratory, Uppsala University in Sweden. The fragment length of the fluorescently labelled fragments was visualized and scored using the software GeneMarker® version 1.6 (Softgenetics). The allele sizes were adjusted to the sizes obtained at SCRI [40]. Mitochondrial DNA (mtDNA) haplotypes of 72 P. infestans isolates were identified by using the method previously described by Griffith [41].
Data analysis
In this study the hypothesis that potato and tomato isolates are equally aggressive in both hosts, potato and tomato detached leaflets (Ho:µPotato isolates=µTomato isolates) was tested with a significance level of (α=0.05). The effects of the isolate origin-host (potato or tomato) combination on aggressiveness components were tested through ANOVA using the GLM procedure of SAS 9.2 (SAS Institute Inc., Cary, NC, USA). The mean separation was done using the statement LSMEANS and the differences between the two groups of isolates (potato or tomato origin) were tested by a t-test procedure (Table 1).
Table 1:Simple sequence repeat (SSRs) multilocus genotypes detected in Phytophthora infestans isolates from Nicaragua collected from July 2009 to January 2010.
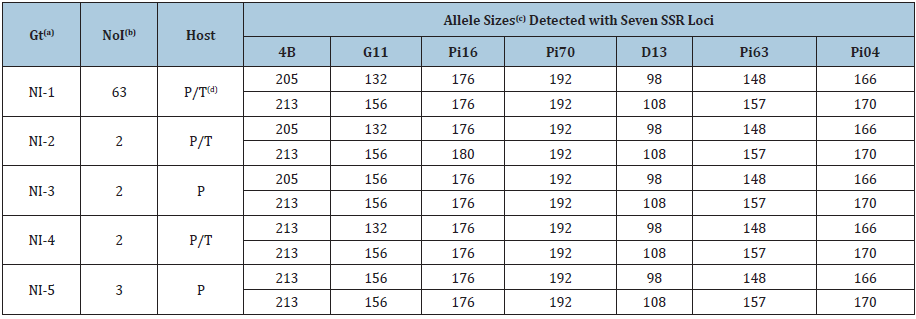
(a)Gt: Genotypes found using seven SSR markers. The genotype NI-1 was the most predominant one, as it was determined in an earlier study addressing genotypic characterization of P. infestans isolates from Nicaragua. The frequency of the other genotypes was very low. (b)NoI: Number of isolates in a given genotype. The NI-1 genotype was common to 46 potato isolates and 17 tomato isolates, whereas 1 potato and 1 tomato isolate shared the same allele sizes and were grouped in the NI-2 and NI-4 genotypes. (c)Allele sizes in bold are indicating where the variation was found. Allele sizes were adjusted to the sizes obtained by Lees et al. [40]. (d)P/T: Potato or tomato host.
Result
Aggressiveness testing
The potato isolate NIC-58 did not grow on potato leaflets, but it grew, although weakly, on tomato leaflets. The tomato isolates 6T, 8T, 9T and NIC-96 did not grow on potato leaflets, but they grew on tomato leaflets (Table 2). A significant effect among potato isolates with regard to IP (P=0.04), LA (P=0.05) and LGR (P=0.01) was found. Potato isolates induced necrotic spots in tomato leaflets earlier than on potato leaflets, produced larger LA in potato leaflets and the LGR was greater in potato leaflets than in tomato ones. Highly significant differences among potato isolates for LP (P<0.0001), SP (P<0.0001), SR (P<0.0001) and AI (P<0.003) were found. Potato isolates had a shorter LP, produced more sporangia (SP), had a greater SR and were more aggressive (AI) on tomato leaflets than in potato ones. Potato isolates were not statistically different regarding SA (P=0.73). No significant differences for IP (P=0.75), LA (P=0.95) and LGR (P=0.33) among tomato isolates were found. Highly significant effects among tomato isolates for LP (P<0.0001), SP (P<0.0001), SA (P<0.006) and AI (P<0.0001) were detected. The mean values for SR among tomato isolates were statistically significant (P=0.05). Tomato isolates had a shorter LP, produced more sporangia (SP), had a greater SA and were more aggressive on tomato leaflets than on potato ones. Potato and tomato isolates both had a shorter LP and higher SP on tomato leaflets compared to potato leaflets. Tomato isolates had the longest LP among tested isolates and that was observed on potato leaflets, whereas potato isolates had the highest SP and that was detected on tomato leaflets. The smallest SA was produced in potato leaflets by tomato isolates. It is noteworthy to point out that, both potato and tomato isolates were more aggressive (AI) in tomato leaflets and produced offspring in greater abundance (SP) on tomato leaflets than in potato ones, i.e., they were prolific in tomato leaflets (Table 3) (Figure 1 & 2).
Table 2:Potato and tomato isolates of Phytophthora infestans from Nicaragua used for determining their aggressiveness on potato and tomato leaflets in cross-inoculation experiments.
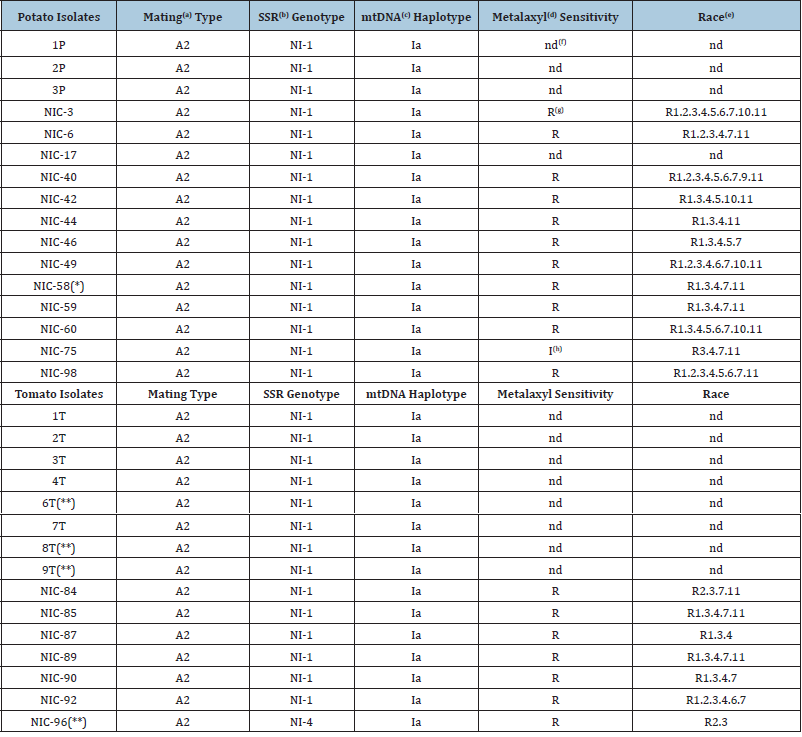
(a)Mating type determination; (b)Simple sequence repeats (also known as microsatellites); and (c)mtDNA haplotyping were done in this study. (d)Metalaxyl sensitivity and (e)race determination was done in a previous study. (f)nd=not determined (unknown). (g)R=isolates resistant to the fungicide metalaxyl. (h)I=isolate with intermediate sensitivity to the fungicide metalaxyl. (*)This isolate did not grow on potato leaflets, albeit weakly it grew on tomato leaflets. (**)These isolates did not grow on potato leaflets, but they grew on tomato leaflets.
Table 3:Least square mean (LSMEAN) values of the aggressiveness components resulting from cross-inoculation tests with potato and tomato isolates of Phytophthora infestans in potato and tomato leaflets.
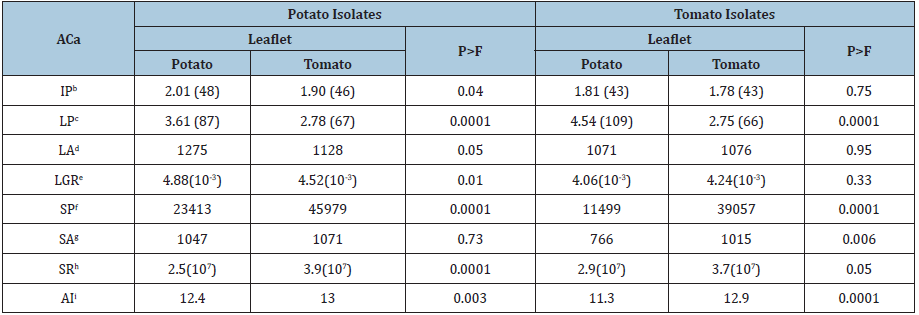
aAggressiveness components bIP=Incubation period [time (days) after inoculation when necrotic spots appeared; in parenthesis is indicated the IP in hours]; cLP=Latency period [time (days) after inoculation when sporangia appeared; in parenthesis is indicated the LP in hours]; dLA=Lesion area (mm2) including the sporulating annulus (divide by 106 to convert it to square meters); eLGR=Lesion growth rate, measured in meters per day; fSP=Spore production calculated by multiplying the sporangia concentration by the volume of a preservative solution [(0.04M copper sulfate, 0.2M sodium acetate/acetic acid, pH 5.4); Mizubuti and Fry [37]]. gSA=Sporulating area (mm2), which is the difference between LA and the area before the LP (hours); divide by 106 to convert it to square meters. hSR=Sporulation rate (sporangia per square meter per day), which is calculated using the equation SR=SP/SA; iAI=Index of aggressiveness, calculated by the formula AI=ln(LA x SP x 1/LP) according to Montarry et al. [38].
Figure 1:Genotypes of Phytophthora infestans detected using simple sequence repeat (SSRs) markers and the percentage of potato (n=53) and tomato (n=19) isolates found in each genotype.
Figure 2:Aggressiveness parameters of P. infestans isolates from Nicaragua measured through cross-inoculation assays using detached potato and tomato leaflets. Vertical bars represent the standard deviation.

SSR fingerprinting and mtDNA haplotyping
Five SSR multilocus genotypes among 72 isolates of P. infestans from Nicaragua were detected and all 72 isolates sampled from potato and tomato fields were of the Ia mtDNA haplotype and A2 mating type. The most predominant was the genotype NI-1, found in 63 out of 72 isolates and reaching a frequency of 87.5%. The NI-1 genotype was common to 46 potato isolates and 17 tomato isolates. The frequency of the remaining four genotypes was very low (Figure 1). Variation in tomato isolates was found only in two isolates at loci 4B and Pi16 and in both cases they shared the same allele sizes with two potato isolates (Table 1). The potato and tomato isolates grouped in NI-2 and NI-4 genotypes were sampled from locations distant from one another 120 (NI-2) and 80 (NI-4) kilometers respectively. In contrast, potato isolates belonging to NI-3 and NI-5 genotypes were sampled from either the same field (NI-3) or neighboring fields (NI-5). In general, two kinds of variations were detected, namely, from heterozygosity to homozygosity at loci 4B and G11 and from homozygosity to heterozygosity at locus Pi16. The common trait of the five identified genotypes is that they belong to the A2 mating type and have the Ia mtDNA haplotype (Table 2). The 4B, G11 and Pi16 loci were the most variable, as they showed differences among tested isolates of P. infestans.
Discussion
This study also undertaken to determine whether there are differences in aggressiveness among potato and tomato isolates through reciprocal aggressiveness tests. A single potato isolates and four tomato isolates did not grow at all on potato leaflets, but they grew on tomato leaflets, indicating that there might be some kind of host preference of these isolates toward tomato. This is in line with findings of other studies in which it has been found host specificity of P. infestans isolates toward either potato or tomato [2,4,27-29]. It is noteworthy to point out that both potato and tomato isolates were kept on their hosts of origin before being used in the aggressiveness tests and the time between subculture of the isolates on potato or tomato leaflets and their use in the tests was relatively short, ten days at the most. Therefore, the reasons behind the loss of pathogenicity of the single potato isolate that could not infect potato leaflets remain unknown.
Some observations done in this study would support the hypothesis that tomato is a better host than potato due to the following:
i. the time elapsed between the end of the IP (appearance of
small necrotic spots) and the beginning of the LP (appearance
of sporangia) was shorter on tomato leaflets than on potato
ones, that is, the LP was shorter than IP, showing that potato
isolates displayed a biotrophic colonization phase on tomato
leaflets as has been reported in other studies [3, 8,42];
ii. the LP was shorter on tomato leaflets than on potato ones;
iii. the LA was greater on tomato leaflets, indicating that
disease intensity is expected to be higher on tomato;
iv. The SP was 1.9 times greater on tomato leaflets than on
potato ones;
v. The aggressiveness index was greater on tomato
leaflets than on potato ones and was almost the same as
the aggressiveness index of the tomato isolates on tomato
leaflets, suggesting that potato and tomato isolates are equally
aggressive on tomato.
Contrary to our initial hypothesis, tomato isolates performed better on tomato leaflets than on potato ones. This finding could be showing host-specificity of tomato isolates toward tomato. In general, tomato isolates were more aggressive on their host of origin, whereas potato isolates were more aggressive on the alternative host.
In a previous work, an SSR fingerprinting analysis showed that Nicaraguan population of P. infestans belonged to a single multilocus genotype referred to as NI-1. Nevertheless, in the same study two rare genotypes that showed one-step allelic differences at the loci Pi16 and G11 were detected (Blandón-Díaz et al., unpublished). In the present study, five SSR multilocus genotypes, four of which had not been previously described, were found among the 72 P. infestans isolates tested. The NI-1 single multilocus genotype continued to be dominant within the clonal lineage, comprising 46 potato isolates and 17 tomato isolates. This finding could indicate that there is neither host specificity nor population differentiation between this group of potato and tomato isolates as has been suggested in an earlier study (Blandón-Díaz et al., unpublished). Moreover, the movement of planting material (infected potato seed tubers and tomato seedlings) among and within production areas seems to be fostering a possible migration (genotype flow) of P. infestans strains between potato and tomato crops and consequently, preventing population differentiation and host specificity. Although at very low frequency, four new genotypes (NI-2, NI-3, NI-4 and NI-5) were detected as a result of variation observed at loci 4B, G11, and Pi16, which were also the most variable loci. Two isolates of NI-2, NI-3 and NI-4 genotypes were detected, and three isolates of NI-5 genotype were found. Two types of variations were observed. The first type of variation seen was in isolates with a loss of heterozygosity at loci 4B and G11. A difference of one allele from these loci that are heterozygous in the commonly found multilocus NI-1 genotype was detected. In a study conducted in Australia with P. cinnamomi, the loss of heterozygosity was attributed to a mitotic crossing over and not to a sexual reproduction or self-fertility [42]. Mitotic recombination was also thought to be responsible for loss of heterozygosity in P. infestans allozymes [43]. The second variation found was a two-step difference from homozygosity (176/176) to heterozygosity (176/180) at locus Pi16 that showed one potato and one tomato isolate. These observed new variants could have arisen by mutation or mitotic recombination as has been stated in previous studies dealing with asexually reproducing populations of P. infestans [8,44] and characterization of the genetic diversity of P. ramorum populations from United States and Europe [45]. In a previous study conducted with P. infestans isolates from Nicaragua, one step difference in allele sizes at loci G11 (from 132/156 to 132/154) and Pi16 (from 176/176 to 174/176) was shown (Blandón-Díaz et al., unpublished). Therefore, the observed differences in microsatellite allele sizes in the present study are in agreement with the microsatellite mutation processes that have been inferred in other studies [46].
The results from the multilocus analysis showed that Nicaraguan population of P. infestans is characteristically clonal in the distribution of genotypic variation, though novel genotypes at very low frequencies were detected. This conclusion is also supported by the predominance of only one mating type (A2). On the one hand, variation at loci 4B and Pi16 led to detection of two novel genotypes (NI-2 and NI-4) until recently unknown that were composed by one isolate of potato and one of tomato each. The isolates belonging to the NI-2 and NI-4 genotypes were sampled from locations that were separated from each other by almost a hundred kilometers. Therefore, a relationship at geographic scale between isolates of these two genotypes was not found. On the other hand, variation found at loci 4B and G11 revealed the presence of the NI-3 and NI-5 genotypes, which were exclusively potato isolates sampled from either the same field (NI-3 genotype) or neighboring potato fields (NI-5). The distribution of genetic variation among these isolates turned out to be found within either a single field or neighboring fields, that is, on a fine geographical scale. No variation was found for mtDNA markers because only the Ia haplotype was detected. Hence, it was confirmed that Ia is still the dominant mtDNA haplotype in Nicaraguan population of P. infestans. This result is in agreement with a previous study (Blandón-Díaz et al., unpublished) in which was suggested that the Ib mtDNA haplotype had been completely replaced by the Ia haplotype. Moreover, it is believed that the Ia haplotype has also replaced the IIb haplotype which was found in herbarium specimen from Nicaragua dating from 1956 [47].
So far, the NI-1 is still the most widely distributed and dominant genotype within the Nicaraguan clonal lineage of P. infestans. It is also known that this genotype is formed by non-host specific potato and tomato strains, which are resistant to metalaxyl and has a complex race structure (Blandón-Díaz et al., unpublished). However, the occurrence of new variants could pose a greater threat for potato and tomato crops in Nicaragua, especially, if these variants are equally or more pathogenic and more ecologically adapted than the predominant NI-1 genotype. Therefore, more extensive sampling at the sites from which isolates were recovered and genotyping of these new variants would be required to track the movement and diversification of these variants. Meaningful shifts in P. infestans populations have been occurred in UK for instance, where in just four years the prevalence of genotype 13_A2 or “Blue 13”, rose to 79% of late blight outbreaks. The “Blue 13” genotype was first detected in the Netherlands in 2004 [48]. A similar situation has been experienced in the United States, where over a period of five years the US-8 genotype became the most widely distributed, dominant and troublesome genotype [10]. Nicaragua imports annually potato seed from the Netherlands, therefore the occurrence of the “Blue 13” genotype in the Netherlands could have very serious epidemiological implications for potato production in Nicaragua regardless of the fact that the “Blue 13” genotype appears to be better adapted to cooler temperatures [49].
Although SSR fingerprinting and mtDNA haplotyping showed no differentiation between potato and tomato isolates, aggressiveness tests revealed that tomato isolates showed a general, but not exclusively, host-specificity and were more aggressive on tomato. In contrast, potato isolates showed host-preference toward tomato detached leaflets and were more aggressive on them. Overall, both potato and tomato isolates were able to attack the alternative host with the abovementioned differences. All of the isolates (including potato and tomato isolates) tested for aggressiveness differentiation belonged to the dominant NI-1 genotype. Only one tomato isolate (NIC-96), grouped in the NI-4 genotype, had a differential host preference, since this isolate grew on tomato leaflets, but it did not grow on potato leaflets. However, other isolates with the NI-4 genotype were isolated from potato. This could mean that the host preference border is very tenuous (at any time the isolate NIC-96 could switch its preference to potato) or that the NI-4 genotype is heterogeneous and contains a mixture of individuals with varying degrees of adaptation.
Conclusion
We would like to thank in particular the invaluable contribution and funding received from the Swedish International Development Agency (SIDA), Swedish Agency for Research Cooperation with Developing Countries (SAREC) and Swedish Institute (SI). We thank Drs Alison Lees and David Cooke (Scotish Crop Research Institute, Dundee) for providing reference isolates of P. infestans. The help of Anna Berlin and Maria Jonsson in planting and care of tomato plants in its early stages in the greenhouse is gratefully acknowledged.
Acknowledgement
We would like to thank in particular the invaluable contribution and funding received from the Swedish International Development Agency (SIDA), Swedish Agency for Research Cooperation with Developing Countries (SAREC) and Swedish Institute (SI). We thank Drs Alison Lees and David Cooke (Scotish Crop Research Institute, Dundee) for providing reference isolates of P. infestans. The help of Anna Berlin and Maria Jonsson in planting and care of tomato plants in its early stages in the greenhouse is gratefully acknowledged.
References
- Haverkort AJ, Boonekamp PM, Hutten R, Jacobsen E, Lotz LAP, et al. (2008) Societal costs of late blight in potato and prospects of durable resistance through cisgenic modification. Potato Research 51: 47-57.
- Legard DE, Lee TY, Fry WE (1995) Pathogenic specialization in Phytophthora infestans: aggressiveness on tomato. Phytopathology 85: 1356-1361.
- Smart CD, Myers KL, Restrepo S, Martin GB, Fry WE (2003) Partial resistance of tomato to Phytophthora infestans is not dependent upon ethylene, jasmonic acid, or salicylic acid signaling pathways. Mol Plant Microbe Interact 16(2): 141-148.
- Vega Sánchez ME, Erselius LJ, Rodriguez AM, Bastidas O, Hohl HR, et al. (2000) Host adaptation to potato and tomato within the US-1 clonal lineage of Phytophthora infestans in Uganda and Kenya. Plant Pathology 49(5): 531-539.
- Fry W (2008) Phytophthora infestans: the plant (and R gene) destroyer. Molecular Plant Pathology 9(3): 385-402.
- Ristaino JB (2002) Tracking historic migrations of the Irish potato famine pathogen, Phytophthora infestans. Microbes and infection 4(13): 1369-1377.
- Fry WE, Goodwin SB (1997b) Resurgence of the Irish potato famine fungus. Bioscience 47(6): 363-371.
- Goodwin SB, Cohen BA, Fry WE (1994) Panglobal distribution of a single clonal lineage of the Irish potato famine fungus. Proc Natl Acad Sci USA 91(24): 11591-11595.
- Goodwin SB, Spielman LJ, Matuszak JM, Bergeron SN, Fry WE (1992) Clonal diversity and genetic differentiation of Phytophthora infestans populations in northern and central Mexico. Phytopathology 82(9): 955-961.
- Fry WE, Goodwin SB (1997a) Re-emergence of potato and tomato late blight in the United States. Plant Disease 81(12): 1349-1357.
- Day JP, Shattock RC (1997) Aggressiveness and other factors relating to displacement of populations of Phytophthora infestans in England and Wales. European Journal of Plant Pathology 103: 379-391.
- Andersson B, Widmark AK, Yuen JE, Nielsen B, Ravnskov S (2009) The role of oospores in the epidemiology of potato late blight. Acta Horticulturae 834: 61-68.
- Hannukkala AO, Kaukoranta T, Lehtinen A, Rahkonen A (2007) Late-blight epidemics on potato in Finland, 1933-2002; increased and earlier occurrence of epidemics associated with climate change lack of rotation. Plant Pathology 56(1): 167-176.
- Kirk WW, Abu El Samen FM, Muhinyuza JB, Hammerschmidt Douches DS, Thill CA, et al. (2005) Evaluation of potato late blight management utilizing host plant resistance and reduced rates and frequencies of fungicides applications. Crop Protection 24(11): 961-970.
- Lehtinen A, Andersson B, Le VH, Nærstad R, Rastas M, et al. (2009) Aggressiveness of Phytophthora infestans on detached potato leaflets in four Nordic countries. Plant Pathology 58(4): 690-702.
- Pilet F, Chacón G, Forbes GA, Andrivon D (2006) Protection of susceptible potato cultivars against late blight in mixtures increases with decreasing disease pressure. Phytopathology 96(7): 777-783.
- Vanderplank JE (1963) Plant diseases: Epidemics and control. Academic Press, New York, USA, p. 349.
- Pariaud B, Ravigné V, Halkett F, Goyeau H, Carlier J, et al. (2009) Aggressiveness and its role in the adaptation of plant pathogens. Plant Pathology 58(3): 409-424.
- Carlisle DJ, Cooke LR, Watson S, Brown AE (2002) Foliar aggressiveness of Northern Ireland isolates of Phytophthora infestans on detached leaflets of three potato cultivars. Plant Pathology 51(4): 424-434.
- Flier WG, Turkensteen LJ (1999) Foliar aggressiveness of Phytophthora infestans in three potato growing regions in the Netherlands. European Journal of Plant Pathology 105: 381-388.
- Montarry J, Glais I, Corbiere R, Andrivon D (2008) Adaptation to the most abundant host genotype in an agricultural plant-pathogen system-potato late blight. J Evol Biol 21(5): 1397-1407.
- Kroon LPNM (2010) The genus Phytophthora; phylogeny, speciation and host specificity. PhD Thesis, Wageningen University, Wageningen, Netherlands, p.184.
- Kamoun S, Smart CD (2005) Late blight of potato and tomato in the genomics era. Plant Disease 89(7): 692-699.
- Adler NE, Chacón G, Flier WG, Forbes GA (2002) The Andean fruit crop, pear melon (Solanum muricatum) is a common host for A1 and A2 strains of Phytophthora infestans in Ecuador. Plant Pathology 51(6): 802.
- Adler NE, Erselius LJ, Chacón MG, Flier WG, Ordoñez ME, et al. (2004) Genetic diversity of Phytophthora infestans sensu lato in Ecuador provides new insight into the origin of this important plant pathogen. Phytopathology 94(2): 154-162.
- Erwin DC, Ribeiro OK (1996) Phytophthora diseases worldwide. The American Phytopathological Society. APS Press, St. Paul Minnesota, USA, p. 562.
- Oyarzun PJ, Pozo A, Ordoñez ME, Doucett K, Forbes GA (1998) Host specificity of Phytophthora infestans on tomato and potato in Ecuador. Phytopathology 88(3): 265-271.
- Suassuna ND, Maffia LA, Mizubuti ESG (2004) Aggressiveness and host specificity of Brazilian isolates of Phytophthora infestans. Plant Pathology 53(4): 405-413.
- Wangsomboondee T, Groves CT, Shoemaker PB, Cubeta MA, Ristaino JB (2002) Phytophthora infestans populations from tomato and potato in North Carolina differ in genetic diversity and structure. Phytopathology 92(11): 1189-1195.
- Goodwin SB, Smart CD, Sandrock RW, Deahl KL, Punja ZK, et al. (1998) Genetic change within populations of Phytophthora infestans in the United States and Canada during 1994 to 1996: role of migration and recombination. Phytopathology 88(9): 939-949.
- Chen CH, Sheu ZM, Wang TC (2008) Host specificity and tomato-related race composition of Phytophthora infestans isolates in Taiwan during 2004 and 2005. Plant Disease 92(5): 751-755.
- Barret LG, Kniskern JM, Bodenhausen N, Zhang W, Bergelson J (2009) Continua of specificity and virulence in plant host-pathogen interactions: causes and consequences. New Phytologist 183(3): 513-529.
- Kawecki T (1998) Red Queen meets Santa Rosalia: arms races and the evolution of host specialization in organisms with parasitic lifestyles. Am Nat 152(4): 635-651.
- Flier WG, Grünwald NJ, Kroon LPNM, Sturbaum AK, van den Bosch TBM, et al. (2003) The population structure of Phytophthora infestans from the Toluca Valley of Central Mexico suggests genetic differentiation between populations from cultivated potato and wild Solanum spp. Phytopathology 93(4): 382-390.
- Lehtinen A, Hannukkala A, Andersson B, Hermansen A, Le VH, et al. (2008) Phenotypic variation in Nordic populations of Phytophthora infestans in 2003. Plant Pathology 57(2): 227-234.
- Andrade Piedra JL, Hijmans RJ, Forbes GA, Fry WE, Nelson RJ (2005) Simulation of potato late blight in the Andes. I: Modification and parameterization of the LATEBLIGHT model. Phytopathology 95(10): 1191-1199.
- Mizubuti ESG, Fry WE (1998) Temperature effects on developmental stages of isolates from three clonal lineages of Phytophthora infestans. Phytopathology 88(8): 837-843.
- Montarry J, Corbiere R, Andrivon D (2007) Is there a trade-off between aggressiveness and overwinter survival in Phytophthora infestans? Functional Ecology 21(3): 603-610.
- Knapova G, Gisi U (2002) Phenotypic and genotypic structure of Phytophthora infestans populations on potato and tomato in France and Switzerland. Plant Pathology 51(5): 641-653.
- Lees AK, Wattier R, Shaw DS, Sullivan L, Williams NA, et al. (2006) Novel microsatellite markers for the analysis of Phytophthora infestans Plant Pathology 55(3): 311-319.
- Griffith GW, Shaw DS (1998) Polymorphisms in Phytophthora infestans: Four mitochondrial haplotypes are detected after PCR amplification of DNA from pure cultures or from host lesions. Appl Environ Microbiol 64(10): 4007-4014.
- Dobrowolski MP, Tommerup IC, Shearer BL, O’Brien PA (2003) Three clonal lineages of Phytophthora cinnamomi in Australia revealed by microsatellites. Phytopathology 93(6): 695-704.
- Goodwin SB (1997) The population genetics of Phytophthora. Phytopathology 87(4): 462-473.
- Abu El Samen FM, Secor GA, Gumestad NC (2003) Variability in virulence among asexual progenies of Phytophthora infestans. Phytopathology 93(3): 293-304.
- Ivors K, Garbelotto M, Vries IDE, Ruyter Spira C, Hekkert BTE, et al. (2006) Microsatellite markers identify three lineages of Phytophthora ramorum in US nurseries, yet single lineages in US forest and European nursery populations. Molecular Ecology 15(6): 1493-1505.
- Goldstein DB, Pollock DD (1997) Launching microsatellites: A review of mutation processes and methods of phylogenetic inference. Journal of Heredity 88(5): 335-342.
- May KJ, Ristaino JB (2004) Identity of the mtDNA haplotype(s) of Phytophthora infestans in historical specimens from the Irish Potato Famine. Mycol Res 108(Pt 5): 471-479.
- Cooke DEL, Lees AK, Shaw DS, Taylor MC, Prentice MWC, et al. (2008) The status of GB blight populations and the threat of oospores. Proceedings Crop Protection in Northern Britain, pp. 217-222.
- Vleeshouwers VGAA, Rietman H, Krenek P, Champouret N, Young C, et al. (2008) Effector genomics accelerates discovery and functional profiling of potato disease resistance and Phytophthora infestans avirulence genes. PLoS ONE 3(8): e2857.
© 2021 Blandón Díaz JU. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






