- Submissions

Full Text
Modern Concepts & Developments in Agronomy
The ALS Gene as Genetic Target in CRISPR/ Cas Approaches: What Have We Learned So Far?
Flavia S Darqui1,2*, H Esteban Hopp3,1 and Marisa López Bilbao1
1Instituto de Agrobiotecnología y Biología Molecular-IABiMo-INTA-CONICET, Instituto de Biotecnología, Centro de Investigaciones en Ciencias Veterinarias y Agronómicas, Instituto Nacional de Tecnología Agropecuaria, Hurlingham, Argentina
2Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), Argentina
3Departamento de Fisiología, Biología Molecular y Celular (FBMC), Facultad de Ciencias Exactas y Naturales, Universidad de Buenos, Ciudad Universitaria, Buenos Aires, Argentina
*Corresponding author: Flavia Soledad Darqui, Instituto de Agrobiotecnología y Biología Molecular-IABiMo-INTA-CONICET, Instituto de Biotecnología, Centro de Investigaciones en Ciencias Veterinarias y Agronómicas, Instituto Nacional de Tecnología Agropecuaria, Nicolás Repetto y De Los Reseros S/Nº (B1686IGC) Hurlingham, Buenos Aires, Argentina
Submission: July 15, 2020Published: August 31, 2020

ISSN 2637-7659Volume7 Issue 2
Abstract
Specific mutations in the conserved domains of the acetolactate synthase (ALS) gene conduct to different key amino acid substitutions that can confer herbicide resistance in different plant species. This outcome has been widely exploited to produce herbicide-resistant agronomic crops as well as to direct many genome editing studies. Therefore, the ALS gene has become a model sequence target to improve our technological skills for more precise CRISPR/Cas nucleotide base substitution in plants, which is essential for modulation/modification of gene function as opposed to the more general gene knock out obtained by indels in conventional genome editing studies. This review summarizes the main knowledge and experiences attained from the use of the ALS gene as a target in CRISPR/Cas studies.
Keywords: Acetolactate synthase; Targeted mutagenesis; Gene targeting; Base editing
Abbreviations: ALS: Acetolactate Synthase; AAS: Amino Acid Substitutions; CBE: cytosine base
Editor; DSBs: Double-Strand Breaks; GE: Genome Editing; HDR: Homology-Directed Repair; NHEJ: Non-Homologous End-Joining; RT: Repair Template; SSN: Sequence-Specific Nuclease
Introduction
Acetolactate synthase (ALS), or acetohydroxyacid synthase (AHAS), is the first enzyme in the pathway for biosynthesis of branched-chain essential amino acids valine, leucine and isoleucine (1,2). Herbicides from the five chemical groups sulfonylurea (SU), imidazolinone (IMI), triazolopyrimidine (TP), pyrimidinyl-thiobenzoates (PTB) and sulfonyl-aminocarbonyl-triazolinone (SCT) inhibit ALS and cause plant death by deprivation of branched chain amino acids. Acetolactate synthase-inhibitor herbicides have been widely used in world agriculture since they were first introduced in 1982 (3). Hence, many crops resistant to ALS-inhibitor herbicides have been commercialized, such as IMI-resistant corn, canola, wheat, rice and sunflower, as well as SU-resistant soybean, sunflower and sorghum (4). However, resistant weeds quickly emerged, i.e. the SU-resistant prickly lettuce identified in 1987 in the United States (5). Since then, many species have evolved resistance to these herbicides globally, because of point mutations in the ALS gene, which produced amino acid substitutions (AAS) in the ALS protein that consequently became less sensitivity to herbicides, but with its intrinsic biological function active (6). Researchers have reported at least 29 AAS endowing herbicide resistance at 8 ALS peptide positions (A122, P197, A205, D376, R377, W574, S653 and G654) in more than 60 species (the amino acid numbering corresponds to the amino acid sequence of ALS in Arabidopsis thaliana). The website http://www.weedscience.org presents an updated record of the resistance patterns acquired by mutant herbicide-resistant weeds to ALS-inhibitors according to each AAS [1]. Studies of gene heritability (7-9) indicated that ALS-associated herbicide resistance is controlled by a nuclear gene with a variable degree of dominance. Generally, in diploid species, resistant ALS alleles are dominant over susceptible wild-type alleles (3).
Genome Editing (GE) comprises different methodologies for genetic modification. One of them is the introduction of targeted DNA double-strand breaks (DSBs) using artificial sequence-specific nucleases (SSNs), such as transcription activator-like effector nucleases (TALENs), zinc finger nucleases (ZFNs) or clustered regulatory interspaced short palindromic repeats (CRISPRs)/Cas9 nucleases. Nuclease-induced DSBs are mainly repaired by two different pathways: non-homologous end-joining (NHEJ) and homology-directed repair (HDR) (10). Double-strand break technologies have two main applications in plant biotechnology. NHEJ-mediated targeted mutagenesis is usually used to generate insertion/deletion (indels) mutations that lead to ORF disruption and gene knockout. On the other hand, HDR-mediated gene targeting is used to introduce desired sequences by homologous recombination between the target locus and a repair template (RT) and this allows both allelic replacement for specific modification of a gene product or site-specific insertion of a sequence.
The CRISPR/Cas system, originally derived from the adaptive immune system of bacteria, has been genetically engineered to function as a robust GE tool in different organisms (11). Its main components are a guide RNA (gRNA) and an associated endonuclease, generally, Cas9 of Streptococcus pyogenes. The gRNA contains a ~20 nucleotide target sequence (protospacer) to direct Cas9 to a specific genomic locus and a scaffolding sequence necessary for Cas9 binding. When the gRNA-Cas9 complex binds to the DNA target, Cas9 generates a DSB upstream of the protospacer adjacent motif (PAM) (11,12), which can be repaired by NHEJ or HDR. A more recently developed GE system, called base editing, generates direct point mutation of a few nucleotides without induction of DSBs, by using a cytosine base (CBE) or an adenine base (ABE) editor (13). CBEs fuse nCas9 (mutant D10A Cas9, with nickase activity) or dCas9 (mutant D10A & H840A Cas9, no endonuclease activity) to a cytidine deaminase (CDA). The CBE, directed by the gRNA to a specific locus, changes C-to-T in a small editing window close to the PAM site. Prior to the use of the CRISPR/Cas technology in plants, researchers have used many GE systems for ALS gene targeting, including TALEN (14) or zinc-finger (15) nucleases, chimeric RNA/DNA oligonucleotides (16-19)or the incorporation of a transfer DNA (T-DNA) harboring a fragment of the herbicide-resistant form of the ALS gene (20-22). As the mechanisms of ALS-associated herbicide resistance have been extensively studied, there is wide scientific background supporting the use of this genetic marker when evaluating different CRISPR/Cas technical approaches. The ALS locus is an ideal target for evaluating different CRISPR/Cas strategies, given the ubiquitous nature of ALS expression and the availability of several ALS-specific point mutations conferring herbicide resistance in a many plant species. Therefore, when CRISPR/Cas reagents are targeted to those specific genetic regions, there are high chances to produce herbicide resistant plants. The induction of certain AAS in the ALS peptide sequence reduces the sensitivity of the ALS enzyme to ALS-inhibitor herbicides but maintaining its intrinsic biological function and implying little or no penalty for plant productivity.
Another feature favoring the selection of the ALS gene in CRISPR/Cas GE strategies is that herbicide selection can facilitate enriching gene editing events and the herbicide resistance phenotype can be visually observed. This allows the detection of mutant events cultured in vitro or in the greenhouse, trough low cost and moderately laborious experiments. The herbicide resistance derived from ALS editing has been used in the direct selection of mutants, in the evaluation of the heritability patterns of CRISPR/Cas-induced mutations and in co-editing strategies. Finally, due to the importance of the ALS gene for plant functionality, its high degree of interspecific genetic conservation would allow researchers to draw some general conclusions from the results obtained in several species. This review is focused in different experimental approaches using the CRISPR/Cas system for GE of the ALS gene in diverse plant species (Table 1) [2-22].
Table 1: Studies applying CRISPR/Cas genome editing in the acetolactate synthase gene.
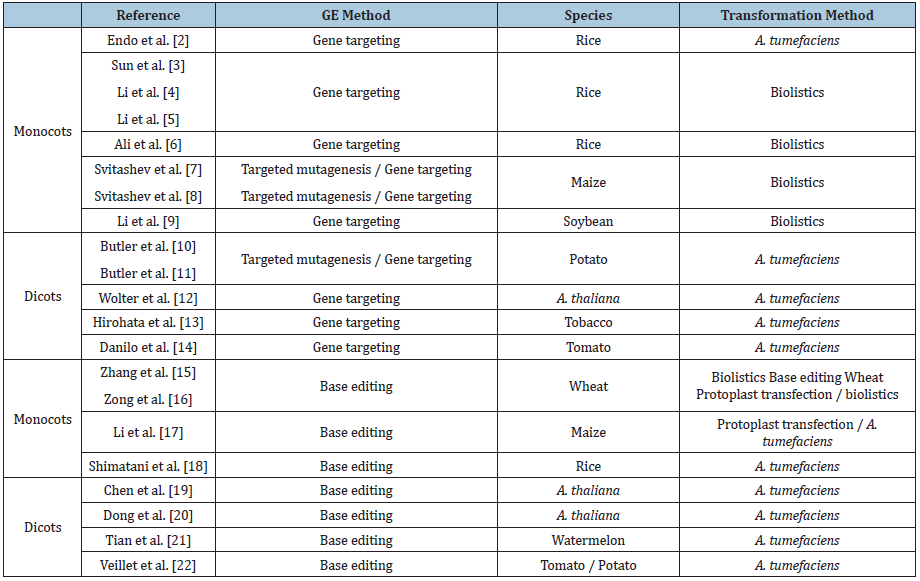
DSB Technologies
Rice
These reports are focused on the gene targeting of the OsALS rice (Oryza sativa) gene and described the use of RTs encoding the W548L and S627I AAS (resistance to bispyribac sodium; PTB) and synonymous mutations to prevent cleavage by the sequence specific endonuclease. Endo et al. [2] synchronized RT delivery and DSB induction by incorporating the CRISPR/Cas reagents through Agrobacterium-mediated rice codon-optimized Cas9 and hygromycin phosphotransferase (p35S::Cas9::tPea3A // 2xp35S::HPT::t35S) and then transformed hygromycin-resistant calli with a second vector harboring two gRNAs and the RT (a partial OsALS sequence). With this strategy, they obtained 0.323% W548L/S627I double mutant callus. To increase HDR efficiency by inhibiting the NHEJ pathway, they added two gRNAs targeting DNA ligase 4 (Lig4) to the Cas9 expression construct. Then, they transformed calli in two steps as previously described. They obtained 0.147 to 1% of double mutant callus. Thus, Lig4 depletion increased editing efficiency. T2 progenies of biallelic gene targeting plants with W548L and S627I mutations in the OsALS gene showed a bispyribac sodium-tolerant phenotype. Another strategy consisted of using a single vector (pOsU3::gRNA1 // pZmUBI::Cas9 // pOsU3::gRNA2 // RT // p35S::HPT) in which the 476-bp RT was flanked with gRNA target sequences so it could be released in vivo thanks to the gRNA-Cas9 complexes [3]. RT availability was ensured by bombarding calli with the vector and free RT at a molar ratio of 1:20. Among 320 bombarded calli, 116 hygromycin and bispyribac sodium-resistant independent lines survived. In addition, from 52 T0 lines randomly selected, 48 were HDR homozygous lines. Edited plants exhibited tolerance to bispyribac sodium. In another approach, Li et al. [4] used a plasmid encoding HPT, a LbCpf1 endonuclease, two crRNAs flanked by ribozymes (RCRs) to facilitate self-processing of mature crRNAS, and a RT with a left homology arm or with two homologous arms. The fact Cpf1 leaves 5' protruding ends after DSB facilitates RT pairing and insertion. Both armed-RTs were flanked with the same two crRNA target sequences to enable the release of the RT from the vector in vivo. Again, RT availability was ensured by bombarding calli with the vector (RT // pZmUBI::LbCpf1 // pOsU3::RCR1::RCR2 // p35S::HPT) and free RT at a molar ratio of 1:20. From 15/152 and 20/164 hygromycin and bispyribac sodium-resistant calli bombarded with the left armed-RT and with the two armed-RT, respectively, they obtained 4 and 7 lines with homologous recombination.
According to the authors, the lower efficiency of OsALS gene replacement achieved using Cpf1 instead of Cas9 [3], may be because of lower editing activity of Cpf1. However, CRISPR/LbCpf1-mediated gene replacement targets sequences that cannot be edited by Cas9 due to differences in PAM requirement. Later, the same group produced OsALS-replacement stable lines by transcript-templated HDR [5]. This approach would ensure RT availability within the nucleus. However, as RT transcripts could be affected by processing and transport to the cytosol, thus leaving RTs unavailable for HDR, the researchers coupled Cpf1 to RCR (crRNAs flanked with ribozymes) units, along with either RDR (RT flanked with ribozymes) or TDT (RT flanked with crRNA targets) units, to produce primary transcripts that self-processed to release the crRNAs and RT inside the nucleus. They tested two strategies, both with a single expression cassette, to ensure that RNA transcripts stay in the nucleus. One strategy consisted of the RCR and RDR units placed in tandem (pOsU3::RCR1::RCR2::RDR::tNOS // pUbi::LbCpf1::tNOS // p35S::HPT) to allow the production of RNA RTs even if the 5ʹ and 3ʹ ends of the primary transcript had been modified.
The other approach coupled two RCR units with a RT flanked with the two crRNAS target sequences (TDT, for target–donor–target) (pOsU3::RCR1::RCR2::TDT::tNOS // pUbi::LbCpf1::tNOS // p35S::HPT) to allow the release of TDT transcripts by the Cpf1-crRNA1 and Cpf1-crRNA2 complexes. From 203 and 193 calli bombarded with RDR and TDT vectors, 19 and 20 calli resulted bispyribac-sodium-resistant, respectively. Further analyses in regenerated plants revealed that HDR editing efficiency was 1.7% (1/58) for the RDR vector and 4.6% (4/87) for the TDT vector. The HDR events presented Mendelian segregation and transgene-free lines could be obtained at T1. Ali et al. [6] used a chimeric protein Cas9-VirD2 and in this way combined the functions of Cas9, which produces targeted DSBs, and of the VirD2 relaxase,, combining the functions of Cas9, which produces targeted DSBs, and the VirD2 relaxase, which brings the RT in close proximity to the DSB site. They introduced pUbi::Cas9, pUbi::Cas9-VirD2 or pUbi::VirD2-Cas9, added to pU6::gRNA-OsALS and an HPT cassette with one of four RT variants: T-RB, T-NRB, mT-RB or mT-NRB. The RTs (T) were designed to include (RB) or exclude (NRB) a 5' right border sequence that would allow covalent binding with VirD2 and a chemical modification of its 5' and 3'ends (m: incorporation of a phosphorothioate linkage) to protect RTs from cellular nucleases. The highest efficiency of HDR editing was obtained in Cas9-VirD2+mT-RB T0 regenerated plants (9.87%) while the efficiency of Cas9+mT-RB plants was 1.56%. This indicated that approximation of the RT to the DSB site increased the repair rate in the replacement of the OsALS wild-type allele by the resistance allele. The results also demonstrated the utility of including the stabilizing modification of the RT and its compatibility with the HDR machinery in vivo, since its incorporation coincided with higher editing rates. The herbicide resistance allele was transmitted to the next generation. Authors mentioned that it would be interesting to explore the fate of the chemically modified RTs in future research, since these chemically modified templates are more stable and have greater chance of genome-wide random insertion. The efficiency parameters obtained in the different works cannot be directly compared, because of the diverse ways of calculating these values. However, considering the number of initial explants and the final number of edited lines obtained as a comparison criterion, the method of Sun et al. [3] seems the most efficient to obtain HDR-edited rice events, so far.
Maize
Svitashev et al. [7] detected that using non-gene-specific gRNAs targeting ALS in mutagenesis experiments caused both ZmALS1 and ZmALS2 maize (Zea mayz) genes to be mutated with similar efficiency, thus affecting the recovery of stable events. Therefore, in subsequent gene targeting experiments, they used a ZmALS2-specific gRNA based on the polymorphisms between ZmALS1 and ZmALS2 nucleotide sequences. They tested three different RTs: a 794-bp fragment of homology cloned into a plasmid vector and two 127-nucleotide single-stranded DNA oligos (Oligo1, Oligo2). All included the P165S modification (resistance to chlorsulfuron; SU) and additional changes to prevent Cas9 cleavage. About 1,000 immature embryos per treatment were bombarded with the two oligo or single plasmid RTs, Cas9, the gRNA targeting ZmALS2 and the MoPAT (maize codon optimized phosphinotricin acetyl transferase)-DsRED gene in DNA expression cassettes and selected for bialaphos resistance. Edited ZmALS2 alleles were detected in two callus sectors from bialaphos-resistant callus sectors generated using the 794-bp RT and in seven callus sectors from chlorsulfuron-resistant callus sectors edited using the 127-nt oligos.
This indicated that small single stranded DNA oligonucleotides were sufficient for gene editing experiments in maize. ZmALS2 editing, using either single-stranded oligos or double-stranded DNA vectors as RTs yielded chlorsulfuron-resistant plants. The evaluated T1 and T2 progeny from two independent T0 plants repaired with the 794-bp fragment and Oligo2 displayed the expected segregation ratio 1:1. Later, the same group co-bombarded embryos with single-stranded oligo as RTs and gRNA-Cas9 RNPs, and selected the co-transformed embryos in chlorsulfuron supplemented medium [8]. Two callus sectors, out of 40 and 50 bombarded embryos, had a mutated and a wild-type allele. Plants regenerated from these callus sectors contained edited ZmALS2 alleles and were chlorsulfuron-resistant. This demonstrated that RNP delivery can enable endogenous gene editing.
Soybean
Li et al. [9] induced the P178S modification (resistance to chlorsulfuron) of the GmALS1 gene (chromosome 4) in soybean (Glycine max). Since there are other three GmALS paralogs in soybean (in chromosomes 6, 13 and 15), they designed a gene-specific gRNA, based on sequence polymorphism around the PAM site with the other GmALS genes. The gRNA-Cas9 vector (pGmU6::gRNA // pEF1A2::Cas9::tPINII) was co-bombarded with free RTs. The RT was a partial GmALS1 mutant sequence that generated the P178S AAS and other silent mutations to prevent the RT from being recognized by the gRNA. Upon direct selection of mutants in chlorsulfuron, one single event was obtained, with both GmALS1 alleles edited, one with the P178S conversion and the other with a 5-base deletion after the DSB site. This demonstrated that precise edition of one of four paralog genes is possible.
Potato
Butler et al. [10] applied NHEJ-targeted mutagenesis in the StALS genes of diploid and tetraploid potato (Solanum tuberosum). Although the two tested gRNAs (gRNA746 and gRNA751) were designed to target StALS1, the paralog StALS2 was also targeted by gRNA751 and contained only a single nucleotide polymorphism in the target site of gRNA746. CRISPR/Cas reagents were delivered by Agrobacterium-mediated transformation by using a conventional expression vector (p35S) or a modified geminivirus expression vector (pLSL). The pLSL vector was co-transformed with another vector coding Rep/RepA (Rep) for replicon release and replication within the plant nucleus. Targeted mutations were detected in calli of both genotypes by using either gRNA in the conventional expression vector. However, mutations were not detected in calli transformed with pLSL. In diploid lines, mutants represented 15% (gRNA746) and 3% (gRNA751) of the lines transformed with the conventional vector and 3% (gRNA746) and 0% of the lines transformed with the geminivirus vector.
In tetraploid lines, mutants were only obtained in the lines transformed with the conventional vector: 29% (gRNA746) and 3% (gRNA751). According to the authors, the lower efficiency of the geminivirus vector to induce NHEJ mutations in both genotypes could be due to a low efficiency of co-transformation along with the vector expressing Rep. Nine diploids and tetraploids T0 mutants derived from the gRNA746 conventional vector were vegetatively propagated for molecular analysis. These plants showed indels ranging from a single bp insertion to a 38 bp deletion. Complete mutagenesis of all StALS alleles was not observed in these events, likely due to ALS being an essential gene. Later, following gene targeting approaches, Butler et al. [11] modified the StALS1 gene using TALEN or CRISPR/Cas nucleases. SSNs reagents were cloned into a conventional (p35S) or modified geminivirus expression (pLSL) vector. Agrobacterium-mediated transformations were conducted in a constitutively expressing Rep mutant. The RT included the W563L and S642T AAS and it was fused with the NPTII gene for kanamycin direct selection of mutant events.
Genetic transformations were conducted with vectors pLSL-TALEN/RT, p35S-TALEN/RT, p35S-TALEN and p35S-CRISPR. The last two mentioned vectors were co-transformed with a modified pLSL vector (pLSLm) which carried the RT but did not include a 35S promoter or SSN reagents. This strategy was used to elucidate if delivering the RT on a geminivirus replicon and the SSNs on a separate vector could improve gene targeting efficiency by altering the coordination of SSN expression and RT availability. From kanamycin-resistant events transformed with pLSL-TALEN/RT and p35S-CRISPR+pLSLm, respectively, 41.7% (5/12) and 12.5% (1/8) were gene targeting events, thus demonstrating the use of geminivirus for delivering GE reagents and a novel approach to gene targeting in potato.
thaliana
Wolter et al. [12] achieved gene targeting in AtALS using egg cell-specific expression of SaCas9 (Staphylococcus aureus Cas9). Their previously developed gene targeting system (34) relied on a stably integrated T-DNA carrying the RT and the Cas9 expression cassette. Cas9 expression led to the simultaneous induction of three DSBs. The RT was excised out of the genome at the same time as a DSB was induced at the target locus to enhance homologous recombination. The expression of Cas9 under a constitutive promoter allowed gene targeting during plant development. Then, gene targeting events transferred in the germline were detected with a frequency of 1/700 by screening seeds or seedlings (35).
By targeting the AtALS gene, Wolter et al. [12] tested new approaches to improve gene targeting frequencies in Arabidopsis. They proved that nCas9 (S. pyogenes) and SaCas9 (S. aureus) were the most efficient nickase and endonuclease enzymes to induce homologous recombination in Arabidopsis. Since their respective gRNAs did not interfere with each other (36), they could be used together to induce double-strand and single-strand break simultaneously in Arabidopsis cells. In this sense, they also evaluated the type of break, both in the target DNA and for the excision of the RT (double-strand and single-strand breaks, and double-strand breaks leaving protruding ends).
Furthermore, they tested whether tissue-specific promoters regulating Cas9 expression could enhance gene targeting efficiency, since there was reported that the expression control of Cas9 by developmentally regulated promoters such as an egg-specific promoter (37) or a reproductive tissue-associated promoter (38) might enhance mutation frequencies in Arabidopsis. They evaluated the use of the constitutive expression promoter PcUbi4-2 (Petroselinum crispum), as well as the developmentally regulated promoters AtCLV3, AtYAO and AtEC1.1/1.2. The RT included the mutation for S653N AAS (resistance to imazapyr; IMI) and silent mutations spanning the gRNA and PAM sequence within the RT. The SaCas9 endonuclease was more efficient than SpCas9 (under the same Ubi promoter) in obtaining gene targeting events (1 out of about 300 instead of 1 out of 700 seedlings). The induction of single-strand breaks in the AtALS gene did not enhance gene targeting efficiency in these experiments. The most efficient strategy was the combination of SaCas9 (DSB in the DNA target and RT) driven by the EC1.1/1.2 promoter: depending on the line, in the very best case 6% of all seeds carried gene targeting events.
Tobacco
Hirohata et al. [13] assessed gene targeting of two tobacco (Nicotiana tabacum) genes: SuRB (ALS) and An2 (MYB transcription factor involved in anthocyanin synthesis). By Agrobacterium-mediated transformation, they incorporated the binary vectors pGII–T1–T2 or pGII–T1–T2–T3, comprising two (T-DNA1- T-DNA2) or three (T-DNA1- T-DNA2 - T-DNA3) independent T-DNAs, respectively. T-DNA1 contained the RT for SuRB: a partial SuRB sequence encoding the W568L AAS (resistance to chlorsulfuron), flanked by the HPT gene and left and right homology arms. T-DNA2 harbored Cas9 and two gRNAs targeting the SuRB and An2 genes (pG10–90::Cas9::trbcSE9 // pAtU626::gRNA1-SuRB // pAtU626::gRNA2-An2). T-DNA3 contained the RT for An2: a partial An2 sequence driven by p35S, with homology arms. Both RTs were designed to prevent the cleavage of Cas9 after gene replacement. Mutant calli were recovered by hygromycin and chlorsulfuron selection. From the 3115 and 4347 explants transformed with pGII–T1–T2 and pGII–T1– T2–T3, respectively, they recovered 16 (0.51%) and 61(1.4%) double-resistant explants.
T-DNA1 integration reached 100% in both pGII–T1–T2 and pGII–T1– T2–T3, whereas T-DNA2 integration was 87.5% for pGII–T1–T2 and 9.8% for pGII–T1– T2–T3. The replacement of SuRB was detected in two T1–T2–T3-derived lines and eight T1–T2-derived lines (four of these eight lines were bi-allelic). All the homologous recombination events occurred across the endogenous SuRB and 5’ homology arm of the randomly integrated T-DNA1. Besides SuRB, the allotetraploid genome of tobacco contains the paralog SuRA gene, which shares 100% sequence similarity with the target region of gRNA1-SuRB. Homologous recombination of SuRA also occurred in one of the T1–T2-derived lines. Even though some T1–T2–T3-derived lines introduced three different T-DNAs and modified the An2 gRNA target site, no signs of homologous recombination in the endogenous An2 were detected. Altogether, co-transformation of multiple T-DNA in a binary vector enabled CRISPR/Cas9-mediated homologous recombination in tobacco.
Tomato
Danilo et al. [14] accomplished gene targeting in the SlALS1 tomato (Solanum lycopersicum) gene by Agrobacterium-mediated transformation of a single expression vector (pUBI::Cas9::tPea3A // pSlU3::gRNA // RT // pNOS::NPTII::tNOS ) which included a 500-bp RT harboring the P186A AAS. They developed a selection protocol for recovery of transgene-free gene targeting events: transformed explants were cultured in kanamycin for a week and then transferred to chlorsulfuron selective medium every 2 weeks. Regeneration events from 37 independent explants produced at least one chlorsulfuron-resistant plant (15% transformation efficiency; 37/244). Molecular assays revealed that 31 events were HDR-mediated edited events (12.7% gene targeting efficiency; 31/244) and that 12 of them (38%; 12/31) were transgene-free. Therefore, the efficiency of T-DNA-free genome editing in the SlALS1 gene in T0 plants was 4.9% (12/244). Even though tomato has three SlALS genes (SlALS1-chromosome 3; SlALS2-chromosome 7; SlALS3-chromosome 6), no off-targeting was detected in SlALS2 or SlALS3 locus in any of the 12 transgene-free events.
The T1 progeny derived from 8 self HDR-mediated edited events (half of them T-DNA-free) were cultured on chlorsulfuron and kanamycin-containing medium for segregation analysis. Although none of the T0 herbicide-resistant plants were homozygous for the mutation, modifications in the SlALS1 gene were transmitted to progeny, therefore yielding homozygous-edited plants. Progenies from T-DNA-free T0 plants were sensitive to kanamycin, whereas T1 plants derived from T-DNA-carrying T0 plants showed segregation for kanamycin resistance. According to the authors, the efficiency of SlALS1 gene editing (12.7%) was high compared with other studies using Agrobacterium-mediated transformation and mentioned in previous paragraphs of this review [1,6,8]. They highlighted that kanamycin selection was important for the isolation of regenerating cells transiently expressing the CRISPR system and that this selection facilitated the detection of transgene-free edited lines, since attempts to select on chlorsulfuron immediately after the 3 days of co-cultivation with Agrobacterium did not produce any herbicide-resistant regenerants. Thus, transient selection on kanamycin may have allowed transfected cells to express Cas9 and gRNA at a level that was enough to ensure efficient DSBs formation and to contain at the same time sufficient RTs to favor HDR of some of these DSBs.
Base Editing
The ever first used CBE system was BE1, which consisted of dCas9 fused to the CDA from rat, APOBEC1. BE1 induced the C-to-T conversion within a deamination window of approximately 5 nucleotides, typically from positions 4 to 8 within the protospacer. Some changes were incorporated to increase base editing efficiency. The editor BE2 fused the C-terminus of dCas9 with the uracil DNA glycosylase inhibitor (UGI) from bacteriophage PBS1, thus preventing the replacement of U by C. Moreover, dCas9 was replaced by nCas9 in BE3, to lower the frequency of indels (≤1%) by avoiding DNA DSB and subsequent NHEJ (41). Another base editing system, the Target-AID (target-activation-induced cytidine deaminase) editor, fused the CDA from Petromyzon marinus (PmCDA1) to nCas9 and featured editing activity in a deamination window of 3-5 nucleotides around position 2 (-18 in the reverse direction) within the protospacer [23]. As far as we know, CBEs used to date in ALS base editing are based on BE3 (43–48) or Target-AID [18,22,24].
Wheat
Zhang et al. [15] produced transgene-free wheat (Triticum aestivum) lines tolerant to nicosulfuron (SU), imazapic (IMI) and quizalofop (ACCase inhibitor) by base editing the TaALS and TaACC genes. For TaALS editing, they used nCas9-PBE (53), a cereal codon-optimized BE3 editor cloned under the maize Ubiquitin-1 promoter. Expression vectors for this CBE (pZmUBI-1::APOBEC1-nCas9-UGI) and a gRNA targeting TaALS-P174 were co-bombarded into immature embryos of Kenong199 or Kenong9204. Among 640 Kenong199-bombarded embryos, 16 (2.5%) T0 plants had base-editing mutations and ten of them were transgene-free. Changes were mostly C-to-T conversions at positions 6, 7 and 8 of the protospacer. The edited plants presented heterozygous, biallelic or homozygous substitutions in 1, 2 or 3 subgenomes, as well as silent mutations. Apart from the expected P174S and P174F mutations, there were also P174A substitutions caused by C-to-G transversions rather than C-to-T transitions, and P174F&R175C double missense mutations caused by additional dual C-to-T transitions at the ninth and tenth position of the spacer sequence.
These 16 base-edited T0 plants with emerging rootlets were transferred to nicosulfuron-supplemented medium. Resistant plants had ≥3 edited alleles, whereas the edited but sensitive plants had one or two missense or silent edits. Transgene-free homozygous mutant T2 plants were exposed at the field-recommended dose of nicosulfuron: whereas wild-type plants did not survive, mutants with 4-6 edited alleles grew normally and mutants with only 2 edited alleles showed different levels of growth retardation.
In genotype Kenong9204, T0 plants regenerated in non-selective medium were subcultured in nicosulfuron-supplemented medium for mutant selection. Ten mutant plants survived, demonstrating that the TaALS-P174 edition generated enough resistance to nicosulfuron to allow the in vitro direct selection of mutants. Later, to evaluate if the nicosulfuron tolerance resulting from TaALS-P174 editing could be used as a co-editing marker, they introduced a second mutation at TaALS-G631 for resistance to imazapic (IMI). About 1200 Kenong199 embryos were co-bombarded with nCas9-PBE and a vector expressing gRNAs targeting TaALS-P174 and TaALS-G631. Thousands of plants regenerated in non-selective medium were transferred to nicosulfuron-supplemented medium and fifty of them survived and grew. All these resistant plants showed multiallelic edits in P174 and 27 (54%) had additional missense edits in the G631-gRNA region. Transgene-free T2 plants with 6 edit alleles in TaALS-P174 and plants with six edit alleles in TaALS-P174+4 alleles edit in TaALS-G631 exposed to imazapic were herbicide tolerant. Moreover, double mutants were 3 to 5 times more tolerant, and were slightly more tolerant to nicosulfuron. According to the authors, this could be due to a synergistic effect of mutations in both regions.
Finally, as mutations in position A1992 of the TaACC gene (acetyl-coenzyme A carboxylase) confer resistance to quizalofop, they coupled TaACCase-A1992 and TaALS-P174 editing to assess if the co-editing system based on TaALS-P174 was efficient in other gene than TaALS. Of the regenerated plants, 22% of nicosulfuron-tolerant plants were edited in TaACCase-A1992. Thus, nicosulfuron tolerance due to AAS in TaALS-P174 was an efficient selection marker for wheat and facilitated the selection of mutants. Furthermore, T2 A1992V transgene-free plants exposed to quizalofop demonstrated that homozygous A1992V mutation in subgenome B conferred plants herbicide resistance. These results confirmed the effectiveness of the TaALS-P174 co-edition strategy, coupling the appropriate gRNAs in the same expression vector. Zong et al. [16] optimized nCas9-PBE by replacing the rat APOBEC1 with plant-codon optimized human APOBEC3A. The A3A-PBE editor was first tested in wheat, rice and potato protoplasts, using gRNAs targeting different genes. An analysis of editing efficiencies at every protospacer position across all target sites revealed that the deamination window for A3A-PBE spanned 17 nt, from protospacer positions 1 to 17 (in comparison to the positions 3 to 9 for nCas9-PBE), and that the frequency of indels was very low.
They also targeted TaALS-P174, aiming to regenerate nicosulfuron-resistant wheat plants. They delivered A3A-PBE and gRNA-TaALS-P174 constructs into 120 immature embryos by particle bombardment and identified 27 mutants harboring at least one C-to-T conversion. Base edits occurred at positions- 7, 6, 7, 8, 9, 10, 12 and 13. Among these 27 mutants, they identified multiple combinations of amino acid substitutions, including 12 mutants with substitutions in all three subgenomes. Outstandingly, six alleles were simultaneously edited in two of these mutants and the deduced proteins all contained amino acid substitutions. The mutant assessed for nicosulfuron resistance was effectively resistant. According to the authors, since ALS genes contain several base-editable codons conferring different herbicide resistances and are conserved across plant species, similar selectable co-editing systems could be readily established to facilitate transgene-free editing with deaminase-Cas9 fusion proteins in other plant species.
Maize
Li et al. [17] produced chlorsulfuron-resistant maize plants using a BE3 editor (pZmU6::gRNA //pZmUBI::APOBEC1::nCas9::UGI // Bar cassette) that targeted P165 in ZmALS1 and ZmALS2. The system was evaluated by protoplast transfection and by Agrobacterium-mediated transformation of immature embryos. Protoplasts showed a C-to-T conversion rate of 2.6% (C7) and 3.4% (C8) in ZmALS1 and 0.6% (C7) and 1.7% (C8) in ZmALS2. T1 plants showed an editing efficiency of 13.9% (16/115) in ZmALS1 with most plants (13; 81%) displaying C7-to-T7 base changes. From these lines, mutant T2 homozygous transgene-free plants were obtained, thus demonstrating the heritability of the mutation. Lines with C7-to-T7 (P165S), C7-to-G7 (P165A) or C7C8-to-T7G8 (P165W) substitutions were herbicide-tolerant. Also, a T3 homozygous double mutant (mutated in both ALS1 and ALS2 genes) was obtained. Transgene-free edited plants harboring an homozygous ZmALS1 mutation or a ZmALS1 and ZmALS2 double mutation were tested and survived at a dose of up to 15-fold the recommended limit of chlorsulfuron. Since the sequence contexts of ZmALS1 and ZmALS2 are very similar, the authors suggested that the bias between the mutation frequencies of both genes was probably due to the chromatin states of their locations. Regarding off-target analysis, 5 putative off-target sites identified in-silico were evaluated and no conversions or indels were detected. In addition, none of the agronomic performance parameters (hundred-kernel weight, plant height and ear height) evaluated in mutants showed significant difference from the wild-type control plants.
Rice
Shimatani et al. [18,24] generated imazamox (IMI)-tolerant plants harboring the A96V AAS, by Agrobacterium-mediated transformation of a Target-AID editor (pOsU6::gRNA // 2xp35S::d/nCas9Os-PmCDA1At::tPea3A // p35S::HPT::tOshsp17.3). Hygromycin-resistant callus lines were transferred to imazamox-supplemented medium. They obtained 3 and 14 resistant lines from dCas9Os-PmCDA1At and nCas9Os-PmCDA1At transformants, respectively. Among the 14 nCas9Os-PmCDA1At tolerant lines, 7 presented the A96V mutation. Editing occurred mostly between positions -19 and -17 (2 and 4 in reverse sense). No off-target mutations were detected. As the T1 progeny of self-pollinated T0 mutants showed independent segregation between the A96V mutation and Cas9 or the selection marker, they confirmed the possibility of obtaining T1 transgene-free A96V mutants. The stable inheritance of mutations was confirmed in T2 seedlings, from the self-pollination of T1 transgene-free plants, which showed imazamox tolerance in in vitro experiments.
They also applied ALS-assisted multiplex targeting to elucidate the function of the OsFTIP1e gene, orthologous to Arabidopsis FTIP1 (predict to regulate long-distance transport of florigen protein components). They induced a Q590X mutation generating a stop codon in the OsFTIP1e coding sequence, by using nCas9Os-PmCDA1At and three gRNAs: two targeting OsFTIP1e-G590 and OsFTIP1e-W483 and another targeting OsALS-A96. Edited calli were selected with hygromycin and imazamox and 168 double-resistant callus lines were obtained. The OsFTIP1e-G590 codon was edited in 144/168 lines (85.7%). Further analyses of some of the mutated lines revealed that 14/37 contained C-to-T mutation resulting in a stop codon at OsFTIP1e-G590, whereas 23/37 contained indels. For OsFTIP1e-W483, 3 point mutations and 10 indels were observed in 13/37 lines. An analysis of co-transmission of mutations in OsFTIP1e and OsALS revealed that 16/37 lines from calli with indels or base substitutions at target sites successfully regenerated into fertile T0 plants. Thus, this strategy generated plants with multiple base substitutions, but marker-free, selectable for their herbicide tolerance. In summary, Shimatani et al. [24] reported the transmission of mutations from the callus to regenerants and their progenies and the generation of selectable marker-free herbicide tolerant rice plants with simultaneous multiplex nucleotide substitutions.
thaliana
Chen et al. [19] obtained tribenuron (SU)-resistant Arabidopsis plants by AtALS -P197 editing. They incorporated the pHEE901 plasmid vector by floral dip. This vector contained a BE3 editor cloned under an egg cell-specific promoter (pU6-26::gRNA::tU6-26// pEC1f::CDA::nCas9::UGI::trbcSE9t // p35S::HPT). Since eggs are the target cells in floral dip, the CRISPR/Cas9 system would express before the first cell division, thus increasing the possibility of obtaining complete homozygous or biallelic mutant plants without mosaicism. Changes of P197 to L, S and F occurred in 4 out of 240 hygromycin-resistant plants, in the form of chimeric, heterozygous and biallelic mutations within the editing window reported by Komor et al. [25]. According to the authors, the low editing efficiency in the first generation (1.7%) could be due to the regulation of BE3 by a transient expression promoter or because of the targeted region, in which nCas9 may be less efficient. Nevertheless, the egg cell-specific promoter-controlled system facilitated edited mutations to be passed to the progeny at high efficiency [26-32].
A high percentage of the progeny from three independent mutant lines resulted tribenuron-resistant: 91%, (42/46), 85.1% (80/94) and 75.8% (94/124). Furthermore, some tribenuron-resistant progeny derived from non-edited plants in the first generation, due to mutations that arose during the second generation.
Later, they obtained imazapic (IMI)-resistant Arabidopsis plants by AtALS-S653 editing (48)[20]. Again, they incorporated the pHEE901 plasmid vector by floral dip method. As G-to-A conversion on AtALS-S653 could confer tolerance to imidazolinone herbicides, they attempted to change C-to-T in the complement strand of S653 codon, targeting the C10 position within the 20 nucleotide protospacer sequence [33-39]. They expected the CBE controlled by an egg cell promoter to re-edit the wild type alleles in egg cells and early embryos. Hence, the diversity of base editing events would increase at later generations, thus allowing the selection of herbicide resistant mutants. Accordingly, they did not detect base edited T1 plants but they identified herbicide-resistant mutants in T3 and T4 generations. Most herbicide resistant plants contained the S653N mutation as a result of G10-to-A10. These results showed that it is possible to obtain imazapic-resistant Arabidopsis plants by using a CBE editor.
Watermelon
Tian et al. [21] produced tribenuron-resistant watermelon (Citrullus lanatus) plants by ALS-P190 editing (P190 corresponding to P197 in A. thaliana) with a vector encoding a BE3 editor (pU6-26::gRNA::tU6-26 // p35S::BE3::tNOS // p35S::BAR::t35S). After Agrobacterium-mediated transformation, they obtained a 23% editing efficiency (45 mutants /199 T0 events). The codon P190 changed to S (17%) and L (6%) and mutations were passed to the next generation. Moreover, non-edited T0 plants originated edited T1 progenies. Besides, non-transgenic T1 mutant plants were recovered. The presence of tribenuron-resistance phenotype was demonstrated in homozygous transgene-free P190S plants. Five regions were identified as potential off target (with ≤5 mismatch to the gRNA target) but none presented editing. Furthermore, no nucleotide changes or indels occurred in the analyzed edited plants. In conclusion, this high-efficient base-editing system generated non-transgenic herbicide-resistant watermelon varieties [40-46].
Tomato and potato
Veillet et al. [22] used a Target-AID editor (pAtU6::gRNA // pUBI::nCas9-PmCDA1 // p35S::NPTII::tNOS) targeting tomato and tetraploid potato ALS genes. As Agrobacterium can be used for transient expression of transcriptional units located on the T-DNA, they developed a selection protocol to obtain T-DNA-free events by transiently expressing the CBE. Three cytidines are present in the edition window of the gRNA sequence: C20, C14 and C13, the last two corresponding to codon CCA in P186. The gRNA targeting P186-SlALS1 was highly similar to the region corresponding to SlALS2, with a single mismatch at position 12. After one or two weeks of kanamycin selection pressure covering the transient expression period of Agrobacterium, tomato plant tissues were transferred to chlorsulfuron selective medium, so that only edited cells could grow and regenerate plantlets, thus simplifying mutant detection among primary transformants [47-49].
Thirty plantlets (12.9%; 30/232) were T-DNA-free. Edition efficiency was analyzed in 105 plants (including the 30 transgene-free plants): 104 displayed mutation(s) at the SlALS1 locus. Up to 28.5% showed indels but 71.4% were base edited. Almost all of these base edits occurred at C14, where any substitution (C-to-T, A or G) is sufficient to change P186 to S, A or W residues, which have been shown to confer chlorsulfuron resistance in tobacco (17). Most were C-to-T changes, some of them being homozygous. No C20-to-T20 homozygous change was found as it would lead to a stop codon (CAA-to-TAA) quite possibly affecting plant regeneration. In general, plants were modified at several C positions. Due to the sequence homology between the gRNA targeting P186-SlALS1 and the SlALS2 gene, they sequenced 51 plants at this locus (including 26 transgenic and 25 T-DNA free genotypes), detecting base editing (37%; 19/51) and indel (16%; 8/51) events. Most base editing events (18/19) were observed at C20 whereas two base conversion events were unexpectedly found at position C24, upstream of the gRNA sequence.
The considerable amount of edition events at SlALS2 locus as compared to SlALS1 target site demonstrated that the off-target potential should be carefully estimated while designing target sequences. Over half (60%) of the T-DNA-free SlALS1 mutants and most (88%) of the transgenic SlALS1 mutants were edited at the SlALS2 locus. In the opinion of the authors, these results suggested that limiting the expression of the CRISPR reagents to a few days reduced the risk of off-target in tomato. Regarding the production of base edited potato lines, 20 plants were regenerated and confirmed to be chlorsulfuron-resistant. All plants harbored mutations in the target sequence. Most mutated plants (75%; 15/20) showed indels in the target site, which likely originated from uracil excision and downstream repair systems. The authors stated this high rate of indels was not surprising considering the number of targeted cytidines in the eight StALS alleles. They also suggested that addition of a uracil DNA glycosylase inhibitor protein (UGI) to the deaminase function may have prevented the formation of indels. The remaining 5 plants were base edited, of which 2 (10%; 2/20) were transgene-free. Base conversion was mainly C-to-G and C-to-T, whereas C-to-A was much less frequent. As previously observed in tomato, base conversion was more frequent in C20 and C14 than in C13. In brief, they efficiently edited the targeted cytidine bases in tomato and potato, therefore obtaining edited but transgene-free chlorsulfuron-resistant plants in the first generation [50-53].
Discussion
As the occurrence of certain point mutations in the ALS gene result in herbicide resistance, these modifications have been widely used not only for obtaining herbicide-resistant crops but also for evaluating different CRISPR/Cas strategies on GE studies. So far, there are few reports on CRISPR/Cas NHEJ-mutagenesis of the ALS gene [6,7,9], most likely due to the essential activity of ALS for plant viability. However, different strategies focused on the ALS gene have been implemented to increase HDR-mediated gene targeting efficiency (Figure 1). Herbicide resistance conferred by site-specific modifications of the ALS gene could be used as a selection marker for direct selection of mutants in targeted mutagenesis [9] and base editing [15] strategies. CRISPR/Cas GE studies demonstrated the possibility to precisely edit one or several genes within the same gene family by an appropriate designing of the gRNA (Figure 2).
Figure 1: Strategies implemented to increase CRISPR/Cas-induced HDR-mediated gene targeting of the acetolactate synthase gene.
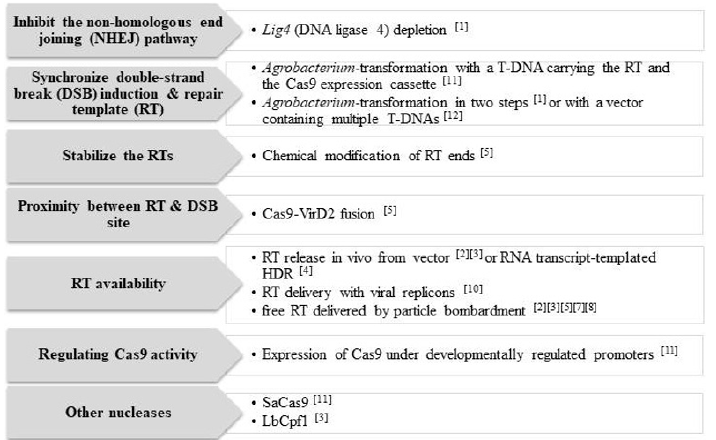
Figure 2: gRNA/s designed for targeting one or several genes within the same gene family. ALS: acetolactate synthase gene.
For example, despite the existence of paralogous genes, researchers could target only one ALS gene in soybean [8] and maize [7]. Otherwise, it was also possible to simultaneously mutate several related genes. One way of multiplexing is to design a single gRNA to target two or more homologous genes sharing a common target site sequence. This outcome was evident in maize [7] and potato [10] NHEJ-targeted mutagenesis. When attempting gene targeting of the SuRB tobacco gene, researchers detected one line with homologous recombination of SuRA, which shared 100% sequence similarity with the target region of gRNA1-SuRB [13]. Also, the use of a single gRNA has been explored in base editing multiplexing approaches, like TaALS editing in the three subgenomes of wheat [15,16] or simultaneous editing of ZmALS1 and ZmALS2 in maize [17]. Multiplexing can also be performed by using two or more gRNAs in a single transformation step in order to edit several unrelated genes at the same time. This method has been successfully applied for ALS-assisted co-editing strategies in maize [15] and rice [18].
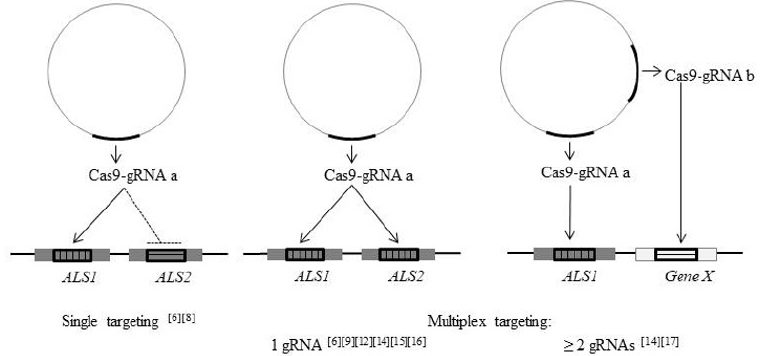
In many plants, the most practical method for T-DNA delivery is Agrobacterium-mediated transformation, since this system can lead longer and more intact DNA with less incorporation of fragmented DNA compared with physical transformation systems. However, simultaneous delivery of Cas9, gRNA(s) and RT(s), along with a selection marker in a single transformation experiment can be complicated, since the efficiency of transformation and integration of a T-DNA decrease as its size increases. In studies focused on HDR-targeted mutagenesis of ALS, CRISPR/Cas reagents delivered by Agrobacterium-mediated transformation have been incorporated in one [12,14] or several T-DNAs [1,13]. Without neglecting that these studies described different gene editing systems being applied in different species, the most efficient strategy seems to be the use of an all-in-one T-DNA, in which the simultaneous incorporation of all CRISPR components within the cell is more assured. The method of particle bombardment, generally used for transformation of recalcitrant species, has been used to ensure sufficient availability of RTs within the cell nucleus [3,4,6,9]. Plant protoplast transfection has been used to assess the functionality of different CBEs in wheat, rice and potato [16] or in maize [17]. Another difficulty for CRISPR/Cas GE is that whenever a T-DNA is delivered into the nucleus, either by biological or physical systems, it will be randomly integrated into the genome, and may produce unwanted side effects such as genetic disruption, mosaicism, etc. In this sense, different options have been evaluated for obtaining transgene-free ALS-edited plants (Figure 3). Main conclusions on the use of the ALS gene in CRISPR/Cas GE studies are summarized in Table 2 [2-8, 10,12-22]. Taken together, all these results allow us to anticipate that the ALS gene will continue to be used as a genetic target in further GE studies and it will remain a valuable tool for greatly expanding our ability to improve agriculturally important traits.
Figure 3: Different approaches for obtaining transgene-free acetolactate synthase (ALS)-edited plants.
Table 1: Main conclusions obtained in different species on the use of the acetolactate synthase (ALS) gene in CRISPR/ Cas genome editing studies.

Acknowledgement
We acknowledge Dr. Julia Verónica Sabio y García for her invaluable assistance with the English text.
References
- Heap I (2020) The international survey of herbicide resistant weeds.
- Endo M, Mikami M, Toki S (2016) Biallelic gene targeting in rice. Plant Physiol 170(2): 667-677.
- Sun Y, Zhang X, Wu C, He Y, Ma Y, et al. (2016) Engineering herbicide-resistant rice plants through CRISPR / Cas9-mediated homologous recombination of acetolactate synthase. Mol Plant 9(4): 628-631.
- Li S, Li J, Zhang J, Du W, Fu J, et al. (2018) Synthesis-dependent repair of Cpf1-induced double strand DNA breaks enables targeted gene replacement in rice. J Exp Bot 69(20): 4715-4721.
- Li S, Li J, He Y, Xu M, Zhang J, et al. (2019) Precise gene replacement in rice by RNA transcript-templated homologous recombination. Nat Biotechnol 37(4): 445-450.
- Ali Z, Shami A, Sedeek K, Kamel R, Alhabsi A, et al. (2020) Fusion of the Cas9 endonuclease and the VirD2 relaxase facilitates homology-directed repair for precise genome engineering in rice. Commun Biol 3(1): 44.
- Svitashev S, Young JK, Schwartz C, Gao H, Falco SC, et al. (2015) Targeted mutagenesis, precise gene editing, and site-specific gene insertion in maize using Cas9 and guide RNA. Plant Physiol 169(2): 931-945.
- Svitashev S, Schwartz C, Lenderts B, Young JK, Cigan AM (2016) Genome editing in maize directed by CRISPR-Cas9 ribonucleoprotein complexes. Nat Commun 7: 13274.
- Li Z, Liu Z, Xing A, Moon BP, Koellhoffer JP, et al. (2015) Cas9-Guide RNA directed genome editing in soybean. Plant Physiol 169(2): 960-970.
- Butler NM, Atkins PA, Voytas DF, Douches DS (2015) Generation and inheritance of targeted mutations in potato (Solanum tuberosum) using the CRISPR/Cas system. PLoS One 10(12): e0144591.
- Butler NM, Baltes NJ, Voytas DF, Douches DS (2016) Geminivirus-mediated genome editing in potato (Solanum tuberosum) using sequence-specific nucleases. Front Plant Sci 7: 1045.
- Wolter F, Klemm J, Puchta H (2018) Efficient in planta gene targeting in Arabidopsis using egg cell-specific expression of the Cas9 nuclease of Staphylococcus aureus. Plant J 94(4): 735-746.
- Hirohata A, Sato I, Kaino K, Iwata Y, Koizumi N, et al. (2019) CRISPR/Cas9-mediated homologous recombination in tobacco. Plant Cell Rep 38(4): 463-473.
- Danilo B, Perrot L, Mara K, Botton E, Nogué F, et al. (2019) Efficient and transgene-free gene targeting using Agrobacterium-mediated delivery of the CRISPR/Cas9 system in tomato. Plant Cell Rep 38(4): 459-462.
- Zhang R, Liu J, Chai Z, Chen S, Bai Y, et al. (2019) Generation of herbicide tolerance traits and a new selectable marker in wheat using base editing. Nat Plants 5: 480-485.
- Zong Y, Song Q, Li C, Jin S, Zhang D, et al. (2018) Efficient C-to-T base editing in plants using a fusion of nCas9 and human APOBEC3A. Nat Biotechnol 36: 950-953.
- Li Y, Zhu J, Wu H, Liu C, Huang C, et al. (2019) Precise base editing of non-allelic acetolactate synthase genes confers sulfonylurea herbicide resistance in maize. Crop J 8(3): 449-456.
- Shimatani Z, Kashojiya S, Takayama M, Terada R, Arazoe T, et al. (2017) Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat Biotechnol 35(5): 441-443.
- Chen Y, Wang Z, Ni H, Xu Y, Chen Q, et al. (2017) CRISPR/Cas9-mediated base-editing system efficiently generates gain-of-function mutations in Arabidopsis. Sci China Life Sci 60(5): 520-523.
- Dong H, Wang D, Bai Z, Yuan Y, Yang W, et al. (2020) Generation of imidazolinone herbicide resistant trait in Arabidopsis. PLoS One 15(5): e0233503.
- Tian S, Jiang L, Cui X, Zhang J, Guo S, et al. (2018) Engineering herbicide-resistant watermelon variety through CRISPR/Cas9-mediated base-editing. Plant Cell Rep 37(9): 1353-1356.
- Veillet F, Perrot L, Chauvin L, Kermarrec MP, Guyon Debast A, et al. (2019) Transgene-free genome editing in tomato and potato plants using agrobacterium-mediated delivery of a CRISPR/Cas9 cytidine base editor. Int J Mol Sci 20(2): 402.
- Nishida K, Arazoe T, Yachie N, Banno S, Kakimoto M, et al. (2016) Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 353(6305): aaf8729.
- Shimatani Z, Fujikura U, Ishii H, Terada R, Nishida K, et al. (2018) Herbicide tolerance-assisted multiplex targeted nucleotide substitution in rice. Data Br 20: 1325-1331.
- Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR (2016) Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533(7603): 420-424.
- Ray TB (1984) Site of action of chlorsulfuron: inhibition of valine and isoleucine biosynthesis in plants. Plant Physiol 75(3): 827-831.
- Duggleby RG, McCourt JA, Guddat LW (2008) Structure and mechanism of inhibition of plant acetohydroxyacid synthase. Plant Physiol Biochem 46(3): 309-324.
- Yu Q, Powles SB (2014) Resistance to AHAS inhibitor herbicides: current understanding. Pest Manag Sci 70(9): 1340-1350.
- Green JM, Owen MD (2011) Herbicide-resistant crops: utilities and limitations for herbicide-resistant weed management. J Agric Food Chem 59(11): 5819-5829.
- Mallory Smith C, Thill D, Dial M (1990) Identification of sulfonylurea herbicide-resistant prickly lettuce (Lactuca serriola). Weed Technol 4(1): 163-168.
- Ashigh J, Rajcan I, Tardif FJ (2008) Genetics of resistance to acetohydroxyacid synthase inhibitors in populations of Eastern Black Nightshade (Solanum Ptychanthum) from Ontario. Weed Sci 56(2): 210-215.
- Scarabel L, Carraro N, Sattin M, Varotto S (2004) Molecular basis and genetic characterisation of evolved resistance to ALS-inhibitors in Papaver rhoeas. Plant Sci 166(3): 703-709.
- Kolkman JM, Slabaugh MB, Bruniard JM, Berry S, Bushman BS, et al. (2004) Acetohydroxyacid synthase mutations conferring resistance to imidazolinone or sulfonylurea herbicides in sunflower. Theor Appl Genet 109(6): 1147-1159.
- Steinert J, Schiml S, Puchta H (2016) Homology-based double-strand break-induced genome engineering in plants. Plant Cell Rep 35(7): 1429-1438.
- Chen K, Wang Y, Zhang R, Zhang H, Gao C (2019) CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu Rev Plant Biol 70(1): 667-697.
- Manghwar H, Lindsey K, Zhang X, Jin S (2019) CRISPR/Cas system: recent advances and future prospects for genome editing. Trends Plant Sci 24(12): 1102-1125.
- Rees HA, Liu DR (2018) Base editing: precision chemistry on the genome and transcriptome of living cells. Nat Rev Genet 19(12): 770-788.
- Zhang Y, Zhang F, Li X, Baller JA, Qi Y, et al. (2013) Transcription activator-like effector nucleases enable efficient plant genome engineering. Plant Physiol 161(1): 20-27.
- Townsend JA, Wright DA, Winfrey RJ, Fu F, Maeder ML, et al. (2009) High-frequency modification of plant genes using engineered zinc-finger nucleases. Nature 459(7245): 442-445.
- Beetham PR, Kipp PB, Sawycky XL, Arntzen CJ, May GD (1999) A tool for functional plant genomics: Chimeric RNA/DNA oligonucleotides cause in vivo gene-specific mutations. Proc Natl Acad Sci U S A 96(15): 8774-8778.
- Kochevenko A, Willmitzer L (2003) Chimeric RNA/DNA oligonucleotide-based site-specific modification of the tobacco acetolactate syntase gene. Plant Physiol 132(1): 174-184.
- Okuzaki A, Toriyama K (2004) Chimeric RNA/DNA oligonucleotide-directed gene targeting in rice. Plant Cell Rep 22(7): 509-512.
- Zhu T, Peterson DJ, Tagliani L, St. Clair G, Baszczynski CL, et al. (1999) Targeted manipulation of maize genes in vivo using chimeric RNA/DNA oligonucleotides. Proc Natl Acad Sci U S A 96(15): 8768-8773.
- Endo M, Osakabe K, Ichikawa H, Toki S (2006) Molecular characterization of true and ectopic gene targeting events at the acetolactate synthase gene in arabidopsis. Plant Cell Physiol 47(3): 372-379.
- Endo M, Osakabe K, Ono K, Handa H, Shimizu T, et al. (2007) Molecular breeding of a novel herbicide-tolerant rice by gene targeting. Plant J 52(1): 157-166.
- Lee KY, Lund P, Lowe K, Dunsmuir P (1990) Homologous recombination in plant cells after agrobacterium-mediated transformation. Plant Cell 2(5): 415-425.
- Fauser F, Roth N, Pacher M, Ilg G, Sánchez Fernández R, et al. (2012) In planta gene targeting. Proc Natl Acad Sci U S A 109(19): 7535-7540.
- Schiml S, Fauser F, Puchta H (2014) The CRISPR/Cas system can be used as nuclease for in planta gene targeting and as paired nickases for directed mutagenesis in Arabidopsis resulting in heritable progeny. Plant J 80(6): 1139-1150.
- Steinert J, Schiml S, Fauser F, Puchta H (2015) Highly efficient heritable plant genome engineering using Cas9 orthologues from Streptococcus thermophilus and Staphylococcus aureus. Plant J 84(6): 1295-1305.
- Wang ZP, Xing HL, Dong L, Zhang HY, Han CY, et al. (2015) Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol 16(1): 144.
- Yan L, Wei S, Wu Y, Hu R, Li H, et al. (2015) High-efficiency genome editing in arabidopsis using YAO Promoter-driven CRISPR/Cas9 system. Mol Plant 8(12): 1820-1823.
- Shimatani Z, Fujikura U, Ishii H, Matsui Y, Suzuki M, et al. (2018) Inheritance of co-edited genes by CRISPR-based targeted nucleotide substitutions in rice. Plant Physiol Biochem 131: 78-83.
- Zong Y, Wang Y, Li C, Zhang R, Chen K, et al. (2017) Precise base editing in rice, wheat and maize with a Cas9- cytidine deaminase fusion. Nat Biotechnol 35(5): 438-440.
© 2020 Flavia S Darqui. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






