- Submissions

Full Text
Modern Concepts & Developments in Agronomy
Detection of Honeybee Viruses in a Queen-Rearing Apiary from Argentina
Ferrufino, Cecilia1; Figini, Emilio2; Gonzalez, Fernanda1; Dus Santos, Maria José1,3 and Salvador, Ricardo4*
1Instituto de Virología, Centro de Investigaciones en Ciencias Agronómicas y Veterinarias (CICVyA), Argentina
2Agencia de Extensión Rural Tandil, Argentina
3Consejo Nacional de Investigaciones Científicas yTécnicas (CONICET), Argentina
4Instituto de Microbiología y Zoología Agrícola (IMyZA). Centro de investigaciones en Ciencias Agronómicas y Veterinarias (CICVyA), Argentina
*Corresponding author: Salvador, Ricardo, Institute of Microbiology and Agricultural Zoology (IMYZA-CICVyA). INTA. Nicolás Repetto and Reseros s/n, 1686 Hurlingham, Provincia de Buenos Aires, Argentina
Submission: February 27, 2020Published: March 17, 2020

ISSN 2637-7659Volume 6 Issue 1
Abstract
Bee viruses cause significant losses in honey production around the world. Previous studies detected at least 8 different viruses in Argentinean honey producer apiaries. Queen rearing activity is used by beekeepers to increase brood and honey production, to start new colonies and to change the colony genetic background. The aim of this study was to detect the presence of bee viruses in an Argentinean queen rearing apiary with highly increased queen pupae mortality. Samples of pupae and royal jelly were collected from queen rearing hives and analyzed by RT-PCR. Sampled pupae with viral symptoms were found to be positive for acute bee paralysis virus (ABPV) and black queen cell virus (BQCV). Gene fragments from ABPV and BQCV positive samples were amplified and sequenced in order to perform phylogenetic analysis. To our knowledge, this is the first report of bee virus detection in a queen rearing apiary from Argentina and the first phylogenetic analysis using local nucleotide sequences data.
Keywords: Argentinean queen rearing apiary; Honeybee; Acute bee paralysis virus; Black queen cell virus; Phylogenetic analysis
Introduction
Argentina is one of the most important honey producers and exporters worldwide. Honeybee (Apis mellifera L.) productivity is threatened by numerous parasites and pathogens such as viruses, bacteria, protozoa, and mites [1]. In addition to parasites and pathogens, loss of queens has been reported as a main reason for colony failure [2]. The presence of a healthy queen is vitally important for colony survival due to several reasons, including her ability to lay large numbers of healthy eggs [3]. Queen rearing apiaries provide young mated queens to beekeepers who need to replace old queens periodically in order to ensure the colony productivity. When replacement activity is performed with infected queens, both vertical and horizontal transmission are enhanced [4]. Bee viruses can exert individual or multiple infections in queen larvae and pupae [5]. In addition, previous reports demonstrated that the virus present in different bee food sources (honey, pollen and royal jelly) may be transmitted horizontally between adult bees such as queens, workers, or drones [6]. Recent studies showed the presence of virus in honeybee colonies from Argentinean producers located along temperate and subtropical regions [7-9]. Notably, many honeybee viruses have been detected in Argentinian stingless bees and bumblebees [10,11]. Black queen cell virus (BCQV), one of the viruses most frequently found in honeybee colonies, is the etiological agent of a lethal disease of honeybee queens [12]. BQCV detection in queen ovaries, eggs and young larvae suggests that vertical transmission from queens to offspring is likely to occur [13].
Recently, BQCV was detected in Argentinean apiaries located along different geographic regions, with a prevalence of 8% [8]. Acute bee paralysis virus (ABPV) is a widespread bee pathogen that has been linked with colony losses [14]. Previous reports demonstrated highly increased percentage of infection by ABPV in Slovenian queen rearing apiaries [5]. ABPV usually shows low prevalence values and its detection is not associated with clinical signs. However, when the virus is present at high titers, elevated mortality rates are evident [14]. ABPV was detected with high prevalence percentage (21.5%) in Argentinean apiaries. In addition to its detection, co-infections with ABPV and BQCV were reported [9]. However, despite increasing knowledge on bee viruses, the information about the presence and effect of virus in Argentinean queen rearing apiaries is lacking. Here, we performed a preliminary study in order to report the presence of bee viruses in one Argentinean queen rearing apiary using RT-PCR and Sanger sequencing. These findings represent the first detection of bee viruses in an Argentinean queen rearing apiary.
Material and Methods
A queen rearing apiary located in Entre Ríos province (Argentina) reported queen pupae mortality that ranged from 40- 100% during last three years. In order to evaluate the presence of ABPV and BQCV viruses, a total of 8 queen pupae samples and 6 royal jelly samples were collected from queen rearing hives (Figure 1). The samples were macerated, and RNA was extracted using the methodology described by Molineri et al. [15].
Figure 1: Viral symptoms observed in the queen rearing process. A. Evidence of the hygienic behavior that allows bees to quickly detect and remove diseased brood; B. A dark inner wall from an affected queen cell (marked with black arrows) compared to a healthy queen cell; C. Cells that were not accepted, 12 cells were accepted from 21 grafted cells; D. Queen cell with dark inner wall.

Virus detection by RT-PCR
Complementary DNA (cDNA) was synthesized by reverse transcription (RT) and PCR was carried out to determine the presence of ABPV and BQCV. The primer sets used were BQCV-F (5’-TGGTCA GCTCCCACTACCTTAAAC-3’), BQCV-R (5’-GCAACAAGAAGAAACGTAAACCAC-3’) [16], and ABPV-F (5′-TCTGATGATGCTGAAGAGAGAAA-3′) and ABPV-R (5′-AATCATCATTGCCGGCTCTA-3) [17] which amplified fragments of 700bp and 500bp respectively. Negative (H2O) and positive controls (recombinant plasmid DNA with the amplicon inserted into the pGEM-T Easy vector) were included in each run of the RT-PCR [13]. The housekeeping gene β-actin was amplified as an internal control using the primers described by Chen et al. [13] to ensure that the entire procedure from RNA extraction to RT-PCR was performed without degradation of RNA.
Phylogenetic analysisSequences were first aligned using Clustal X [18] and phylogenetic trees were inferred using MEGA 7 program with default parameters using Neighbor-Joining (NJ) method and Bootstrap with 1,000 replicates.
Result and Discussion
Viruses ABPV and BQCV were detected in 62.5% (5/8) and 50% (4/8) of queen pupae samples, respectively (Figure 1). Co-infections with both viruses were observed in 37.5% (3/8) of queen pupae samples. ABPV was also detected in 33% (2/6) of the royal jelly samples suggesting a putative source of virus that could infect pupae (Figure 2A&2B). In accordance with these results, previous studies have demonstrated the presence of bee viruses in royal jelly [6]. However, the importance of this food source in viral transmission needs to be further studied. In order to assess the genetic relationship between ABPV and BQCV detected in Argentinean queen rearing apiaries with strains of various geographic origins, a phylogenetic analysis was performed. BQCV and ABPV amplicons from positive pupae samples were sequenced using the primer sets described above. Both, ABPV and BQCV fragments (500bp and 700bp, respectively) belonged to the coding region of their respective structural proteins (VP). BQCV (GenBank MH053385) and ABPV (GenBank MH077964) sequences obtained from positive samples were used to construct phylogenetic trees including reference strains retrieved from the GenBank database (Figure 3).
Figure 2: Viruses’ detection. Viral RNA was obtained from queen pupae and royal jelly. RT-PCR was conducted using specific primers that amplify sequences from ABPV and BQCV. A. ABPV RT-PCR. Pupae samples (1-8); 100 base pairs (bp) DNA Ladder (9); Negative control (10); Positive control (11); Royal jelly samples (12-17). B. BQCV RT-PCR: Pupae samples (1-8); 100 bp DNA Ladder (9); Negative control (10); Positive control (11).
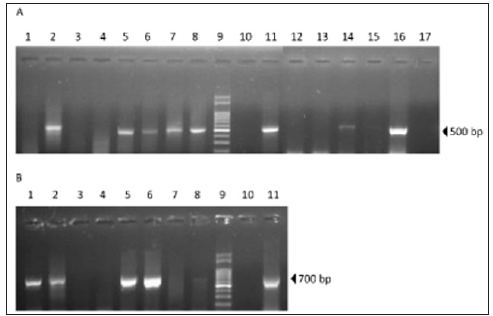
Figure 3: Phylogenetic analysis. A. Neighbor joining tree of 500 bp fragment gene that encodes for a structural protein of ABPV. GenBank accession numbers are indicated in parentheses: Hungary (AY053374.1, AY053372.1 and AY053375.1), Poland (AY053371.1 and AY053370.1), Austria (AY053366.1), Germany (AY053367 and AY053368.1), South Africa (AF150629.1), USA (HM228893.1 and HM228890.1), Brazil (EU292210.1), Uruguay (AY763414.1) and Chile (KF011920.1). 1000 bootstrap replicates were used. Bootstraps values are shown above the branches. B. Neighbor joining tree of 700bp fragment gene that encodes a capsid polyprotein of BQCV. GenBank accession numbers are indicated in parentheses: Switzerland (HG779864.1), China (JX679492.1 and JN185926.1), South Korea (EU375537.1), Taiwan (GU108221.1), Hungary (EF517515.1), Peru (HG974555.1), England (GU903464.1), USA (HQ655458.1), Uruguay (DQ364629.1), South Africa (AF183905.1), Germany (AF521640.1), Chile (KF011921.1) and Brazil (EU292211.1). 1000 bootstrap replicates were used. Bootstraps values are shown above the branches.
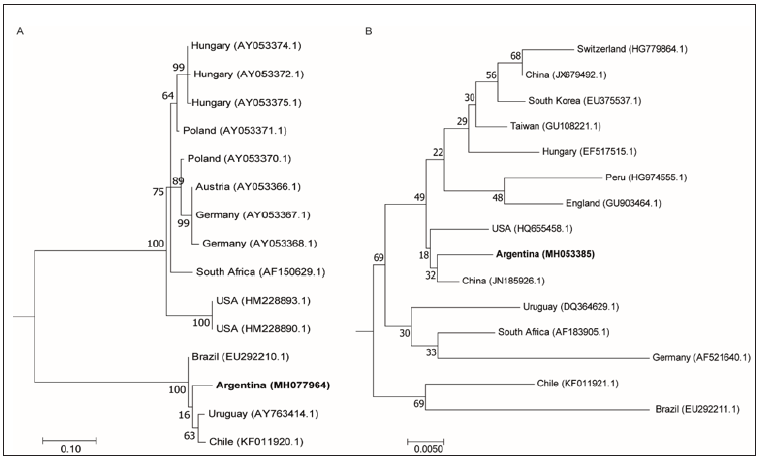
The results showed that ABPV sequences from Argentina, Brazil, Uruguay and Chile are closely related into a South American group that diverged from the European, South Africa and US groups (Figure 3A). The proximity of South American ABPV sequences suggests a possible common origin. Previous studies demonstrated that ABPV genetic diversity support the concept of geographical segregation of bee virus strains [19]. Interestingly, the analysis of BQCV sequences did not show geographical grouping. Thus, we found that Argentinean sequence grouped closer to China and US sequences than to South American ones (Figure 3B). Unexpected grouping in BQCV phylogenetic analysis were previously reported [20]. The genetic recombination between BQCV genotypes and the global apicultural trade could explain the close genetic distance of virus isolates that are geographically unrelated [21,22]. More viral sequences are needed to confirm the results obtained regarding the phylogenetic origin of Argentinean viruses.
Conclusion
In summary, this is the first report describing the presence of bee viruses in a queen rearing apiary in Argentina. This work also shows the first phylogenetic analysis that includes Argentinean bee virus sequences. Further studies are needed in order to determine the prevalence and impact of viral infections on queen rearing process.
Statement of Author Contribution
Conceived and designed the experiments: CF, MJDS and RS. Performed the experiments: CF, EF, FG and RS. Analyzed the data: CF, MJDS and RS. Wrote the manuscript: CF, MJDS and RS. Contributed to the critical revision of the manuscript: CF, EF, FG, MJDS and RS.
Funding
This work was supported by the Instituto Nacional de Tecnología Agropecuaria (INTA). The Cecilia Ferrufino´s grant was supported by the Instituto Nacional de Tecnología Agropecuaria.
Conflict of Interest
The authors declare that they have no conflict of interest.
References
- Tantillo G, Bottaro M, Di Pinto A, Martella V, Di Pinto P, et al. (2015) Virus infections of honeybees Apis Mellifera. Ital J Food Saf 4(3): 5364.
- Genersch E (2010) Honeybee pathology: current threats to honeybees and beekeeping. Appl Microbiol Biotechnol 87(1): 87-97.
- Tarpy D, Seeley D (2006) Lower disease infections in honeybee (Apis mellifera) colonies headed by polyandrous vs monandrous queens. Naturwissenschaften 93(4): 195-199.
- Amiri E, Strand MK, Rueppell O, Tarpy DR (2017) Queen quality and the impact of honeybee diseases on queen health: Potential for interactions between two major threats to colony health. Insects 8 (2).
- Gregorc A, Bakonyi T (2012) Viral infections in queen bees (Apis mellifera carnica) from rearing apiaries. Acta Vet Brno 81(1): 15-19.
- Shen M, Cui L, Ostiguy N, Cox Foster D (2005) Intricate transmission routes and interactions between picorna-like viruses (Kashmir bee virus and sacbrood virus) with the honeybee host and the parasitic varroa mite. J Gen Virol 86(Pt 8): 2281-2289.
- Reynaldi FJ, Sguazza GH, Pecoraro MR, Tizzano MA, Galosi CM (2010) First report of viral infections that affect argentine honeybees. Environ Microbiol Rep 2(6): 749-751.
- Ding G, Fondevila N, Palacio MA, Merke J, Martinez A, et al., (2016) Prevalence of honeybee viruses in different regions of China and Argentina. Rev Sci Tech 35(3): 825-833.
- Molineri AI, Pacini A, Giacobino A, Bulacio Cagnolo N, Aignasse A, et al. (2017a) Prevalence of honeybee (Apis mellifera) viruses in temperate and subtropical regions from Argentina. Rev Argent Microbiol 49(2): 166-173.
- Alvarez LJ, Reynaldi FJ, Ramello PJ, Garcia MLG, Sguazza GH, et al. (2018) Detection of honeybee viruses in Argentinian stingless bees (Hymenoptera: Apidae). Insect Soc 65(1): 191-197.
- Reynaldi FJ (2013) Co-infection of honeybee viruses in asymptomatic Bombus atratus of Argentina. Brazilian Journal of Biology 73: 797-800.
- Bailey L, Woods RD (1977) Two more small RNA viruses from honeybees and further observations on sacbrood and acute bee-paralysis viruses. J Gen Virol 37(1): 175-186.
- Chen YP, Pettis JS, Collins A, Feldlaufer MF (2006) Prevalence and transmission of honeybee viruses. Appl Environ Microbiol 72(1): 606-611.
- De Miranda JR, Cordoni G, Budge G (2010) The Acute bee paralysis virus-Kashmir bee virus-Israeli acute paralysis virus complex. J Invertebr Pathol 103 Suppl 1: S30-47.
- Molineri A, Giacobino A, Pacini A, Bulacio Cagnolo N, Fondevila N, et al. (2017b) Risk factors for the presence of Deformed wing virus and Acute bee paralysis virus under temperate and subtropical climate in Argentinian bee colonies. Prev Vet Med 140: 106-115.
- Benjeddou M, Leat N, Allsopp M, Davison S (2001) Detection of acute bee paralysis virus and black queen cell virus from honeybees by reverse transcriptase pcr. Appl Environ Microbiol 67(5): 2384-2387.
- Teixeira EW, Chen Y, Message D, Pettis J, Evans JD (2008) Virus infections in Brazilian honeybees. J Invertebr Pathol 99(1): 117-119.
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25(24): 4876-4882.
- Bakonyi T, Grabensteiner E, Kolodziejek J, Rusvai M, Topolska G, et al. (2002) Phylogenetic analysis of acute bee paralysis virus strains. Appl Environ Microbiol 68(12): 6446-6450.
- Noh JH, Reddy KE, Choe SE, Yoo MS, Doan HT, et al. (2013) Phylogenetic analysis of black queen cell virus genotypes in South Korea. Virus Genes 46(2): 362-368.
- Tapaszti Z, Forgách P, Kovágó C, Topolska G, Nowotny N, et al. (2009) Genetic analysis and phylogenetic comparison of Black queen cell virus genotypes. Vet Microbiol 139(3-4): 227-234.
- Tsevegmid K, Neumann P, Yanez O (2016) The honeybee pathosphere of mongolia: European viruses in Central Asia. PLoS One 11(3): e0151164.
© 2020 Salvador, Ricardo. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






