- Submissions

Full Text
Modern Concepts & Developments in Agronomy
Interaction of Host Genotype, Medium of Inoculum, and Pathogen Species on Pathogenicity of Pythium Disease on Soybean
Feng Lin1†, Yingdong Bi1,2†, Xuan Gao1,3, Cuihua Gu1, Zixiang Wen1, Janette Jacobs1, Martin I Chilvers1 and Dechun Wang1*
1Department of Plant, Soil and Microbial Sciences, Michigan State University, USA
2Heilongjiang Academy of Agricultural Sciences, China
3Anhui Provincial Key Laboratory of the Conservation and Exploitation of Biological Resources, Anhui Normal University, China
*Corresponding author: Dechun Wang, Department of Plant, Soil and Microbial Sciences, Michigan State University, 1066 Bogue St., Rm. A384-E, East Lansing, MI 48824-1325, USA
Submission: February 04, 2020;Published: February 10, 2020

ISSN 2637-7659Volume5 Issue5
Abstract
Soybean (Glycine max (L.) Merr.) is an important crop for edible oil and protein in the United States and the world. However, soybean yield productions have been challenged by diseases caused by soil-borne pathogens such as Pythium. The purpose of this case study was to identify the interactions among soybean genotypes, medium of inoculum, and pathogen species on the pathogenicity of Pythium disease on soybean. Three-way ANOVA indicated that the interaction of host genotype, inoculum medium and pathogen species significantly affected the severity of Pythium disease on soybean. There were also significant interactions between inoculum medium and pathogen species. Marginal analysis for inoculum medium indicated that rice may be a better medium used for inoculation for Pythium than millet as it induces higher pathogenicity on soybean plants. Marginal analysis for Pythium species indicated that P. oopapillum was the most aggressive on 6 soybean genotypes evaluated by RR and on 4 genotypes by RW; P. irregulare was the most aggressive on 2 soybean genotypes evaluated by RR, and on 4 genotypes by RW; and P. sylvaticum was the most aggressive on one soybean genotype by both RR or RW.
Keywords: Soybean; Pythium root rot; Medium
Abbreviations: ANOVA: Analysis of variance; RR: Root Rot Rating; RW: Ratio of Plant Fresh Weight
Introduction
Soybean (Glycine max (L.) Merr) is one of the most important crops for edible oil and proteins in the United States. However, about 11% of soybean production is suppressed by soybean diseases including seedling disease [1,2]. In the U.S. and Ontario, Canada, seedling diseases caused a total yield loss of 241 million bushels from 2010 to 2014, being the second most destructive soybean diseases [1]. One of the most important pathogens causing soybean seedling diseases is Pythium, a genus of soil-borne, oomycete pathogens typically favored by cool, wet conditions and early planting, causing seed decay, root rot, and damping off [3]. More than 20 Pythium species have been identified with pathogenicity on soybean so far, such as P. ultimum, P. aphanidermatum, P. debaryanum, P. irregulare, P. myriotylum, P. torulosum and P. vexans, in Ohio, Iowa, Florida, Arkansas, Illinois, and North Dakota [4-12]. All the Pythium species cause similar symptoms, including soft and rotted seeds before germination, pre- or post-emergence damping-off, and hypocotyl or root discoloration [13,14]. Crop rotation is a traditional way to manage Pythium diseases [15], however, because Pythium has multiple hosts such as corn and wheat, crop rotation is considered less effective [13]. Another widely used method is the treatment of seeds with fungicide, such as metalaxyl, mefenoxam and some newer fungicides [16-18]
However, the continued use of these fungicides may lead to reduced efficacy due to increased insensitivity of the Pythium population. In addition, fungicide has been more and more widely considered as less desirable for environment and health concerns [19]. The most cost-effective and environmental-friendly method to control disease is the deployment of resistant soybean varieties. To evaluate the soybean seedlings at early stage, greenhouse essay has been developed and used in several studies and rice and millet are the most used medium for inoculum [20-29]. However, it has not been clear yet about which medium is more suitable to evaluate the pathogenicity of Pythium on soybean, and moreover, it is still unknown whether there’s any interactions between soybean genotypes, the inoculum medium, and different Pythium species, which would strongly impact our strategy of evaluating Pythium diseases.
Therefore, this case study was designed to
1) explore the interactions of soybean genotype, inoculum medium, and Pythium species;
2) identify a more suitable medium for Pythium disease evaluation; and
3) Compare the pathogenicity of different Pythium species
Case Presentation
A total of 9 Michigan State University improved soybean lines were used in this study. For the pathogen, three Pythium isolates were used including CMISO2 5-14, MSIO 8-23, and CMISO2 2-30, which were classified as P. irregulare, P. oopapillum, and P. sylvaticum, respectively [3]. All isolates were maintained on slants of Potato Carrot Agar (PCA) at 15 °C in glass tube. A modified protocol from Howell [30] was used to prepare inoculum (Figure 1A). Two cultural media, millet and white rice, were used. 625g millet seeds and 425g white rice (parboiled) grains were each mixed with 400mL ddH2O in an autoclave-safe plastic bag and autoclaved at 121 °C for 4 hours. When bags cooled down to room temperature, one full plate of a 4-day-old isolate cultured at CMA medium was transferred to the millet or rice bag aseptically. After incubation at room temperature for 12 days, the inoculum was ready to use (Figure 1B). The disease was evaluated at the Michigan State University greenhouse, with temperature at 20-22 °C and humidity at 20%. All soybean varieties were evaluated in a randomized incomplete block design with 12 seeds evaluated for each replicate. Three replicates were performed for inoculation group, and one replicate was used as control. To start the assay, 6-cell seed starting trays were first fulfilled with medium size vermiculite and soaked in water until the vermiculite was fully saturated. Then two 2cm-depth holes were made in each cell and approximately 2g inoculum and un-inoculated medium (or 6-8 rice grains or 15-20 millet grains) was placed at the bottom of each hole for inoculation. A single seed was then planted on the top of the inoculum in each hole and covered with vermiculite (Figure 1C). After planting, seed starting trays were transferred to the greenhouse benches which had been covered with waterproof plastic. Benches were watered until the water height reached the height of inoculum. After that, benches were watered every other day to maintain the water saturation environment until two days before measurement. Data were collected when the control group reached V2 stage, or 14 days after inoculation (Figure 1D). Root score and whole plant weight were measured. To rate root score, roots were removed from the pots, gently washed and cleaned with running tap water, and wrapped in a towel to absorb excess water.
Figure 1: Methods used to evaluate soybean partial resistance to Pythium irregulare. A: Growth of P. irregulare isolate (CMISO2 5-14) on the surface of corn meal agar (CMA) on the fourth day. B: Pythium inoculums produced on millet (upper) and rice (lower) media for twelve days. C: Planting of seeds and inoculum in half-inch deep holes. D: Example of inoculated and control plants 14 days post inoculation (dpi). E: Response of soybean plants to P. irregulare 14 dpi. Numbers beneath each plant indicates the root rot (RR) scores with 5 for kill, and 0 for complete health.

Root rot score for each plant was rated on a 0-to-5 scale (Figure 1E), wherein, 0 represents a healthy root system with no symptoms of lesions or rot on the roots; 1 represents small lesions and discoloration on the roots, with approximately 1% to 20% of roots showing visible symptoms; 2 represents some lesions on roots and lateral roots with approximately 20 to 40% of roots rotted with visible symptoms of lateral roots reduction; 3 represents rot on lateral roots and visible symptoms of rot, with approximately 40 to 60% of the roots exhibiting visible symptoms; 4 represents both lateral roots and main tap roots having visible symptoms on roots and approximately 60 to 90% of the roots infected, roots and lateral roots reduced drastically, and 5 represents roots severely rotted or no germination with complete colonization on the seed. Whole plant weight was measured using a bulk of 12 plants for each replicate using an electronic balance (Scout Pro, SP 4001; Ohaus Corp, Pine Brook, NJ). To evaluate the disease, root rot rating (RR) and ratio of fresh plant weight (RW) were used. Root rot rating (RR) was calculated as ∑[5×(number of roots at 5) + 4×(No. of roots at 4) + 3×(No. of roots at 3) + 2×(No. of roots at 2) + 1×(No. of roots at 1) + 0×(No. of roots at 0) ]/[5× (No. of control roots) ]; Ratio of plant fresh weight (RW) was as [Fresh plant weight of inoculated plants / (No. of inoculated seeds] / [Fresh plant weight of control plants / (No. of control plants)]. The analysis of variance (ANOVA) for RR and RW were analyzed using the general linear model procedure (GLM) of SPSS 22 (SAS Institute Inc.) and the results were considered significant with p<0.05. Discrimination among the means was conducted using Fisher’s least significant difference (LSD) procedure.
Figure 2: Comparison of inoculum medium on the pathogenicity of three Pythium species on nine soybean genotypes. A: Evaluation of the root rot rating (RR) for millet (blue) and rice (green). B: Evaluation of the ratio of plant weight (RW) for millet (blue) and rice (green).
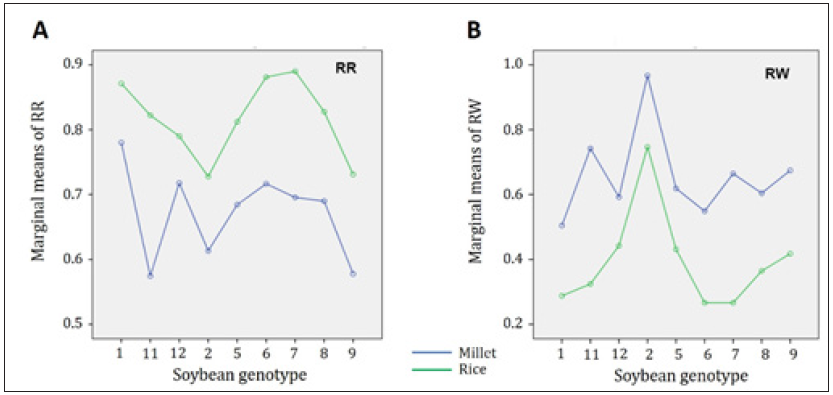
Table 1: Three-way ANOVA for interaction of soybean genotype, inoculum medium, and pathogen species on the pathogenicity of Pythium diseases on soybean plants, evaluated by root rot rating (RR) and ratio of plant fresh weight (RW).
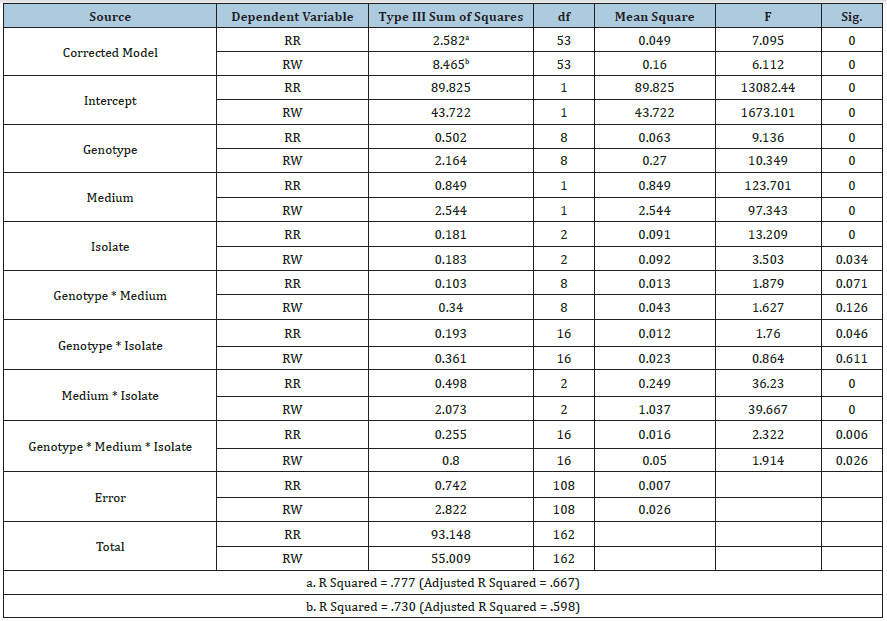
Figure 3: Comparison of the pathogenicity of three Pythium species on nine soybean genotypes. A: Evaluation of the root rot rating (RR) for three Pythium species. B: Evaluation of the ratio of plant weight (RW) for three Pythium species
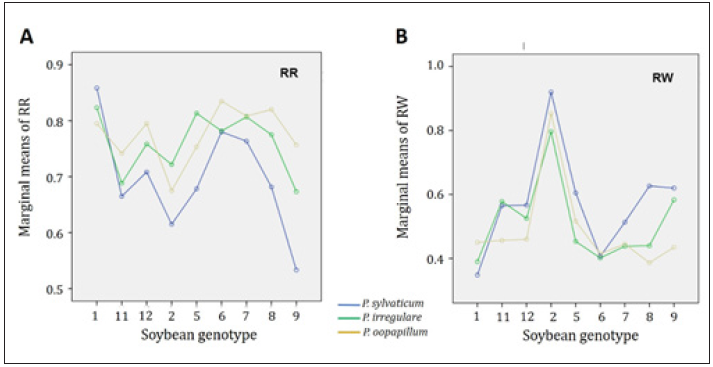
Additionally, we also conducted the marginal analysis to compare the pathogenicity among the three isolates from the three Pythium species (Figure 3). The pathogenicity of three isolates was generally consistent on soybean genotypes using either RR or RW as evaluation method. For example, for soybean genotype1, CMISO2 2-30 (P. sylvaticum) was the most pathogenic as indicated by both RR and RW, followed by CMISO2 5-14 (P. irregulare) and MISO 8-23 (P. oopapillum), while for soybean genotype 9, the most aggressive isolate was the P. oopapillum. The P. oopapillum isolate was most aggressive on six soybean genotypes evaluated by RR (11,12,6,7,8 and 9) and on four soybean genotypes by RW (11,12,8 and 9). The P. irregulare isolate was most aggressive on two soybean genotypes (2 and 5) by RR, and on four genotypes (2,5,6, and 7) by RW. The P. sylvaticum isolate was the most aggressive only on soybean genotype 1.
The different response pattern of soybean genotypes to different species of Pythium suggested that soybean does not deploy a single genetic resistance mechanism against different Pythium species. This can be supported by the identification of different quantitative disease resistance loci (QDRL) for different types of Pythium species. For example, our previous study identified two QDRL on soybean chromosomes 11 and 20, respectively, for partial resistance to P. irregulare [23], while in our recent study for soybean resistance to P. sylvaticum, novel QDRLs were identified on chromosomes 10 and 18 (unpublished data). Nevertheless, some common genes may play critical roles in regulating soybean resistance to different Pythium species. For example, the QDRL on chromosome 20 was identified for both P. irregulare and P. sylvaticum (unpublished data).
Summary
This case study illustrated that soybean genotypes, inoculum media, and types of Pythium isolates, along with the interactions between inoculum media and types of Pythium isolates, and the interactions among the three factors, can significantly affect the soybean response to Pythium pathogen which was evaluated by RR and RW. Compared to millet medium, rice medium can induce heavier disease severity and may serve as a better choice for soybean disease resistance studies for Pythium root rot. In addition, this case study indicated that P. oopapillum was the most aggressive on 6 soybean genotypes evaluated by RR and on 4 genotypes by RW; P. irregulare was the most aggressive on 2 soybean genotypes evaluated by RR, and on 4 genotypes by RW; and P. sylvaticum was the most aggressive on one soybean genotype. Moreover, our results suggested that different mechanisms of resistance may be deployed by soybean plants although some common genes may play important roles for resistance to different Pythium species.
Acknowledgement
The authors appreciate Dr. Shabir H Wani and Dr. Na Zhang for valuable suggestions of the manuscript. This study is supported by Michigan Soybean Promotion Committee, USDA National Institute of Food and Agriculture (Hatch project 1011788) and Ag Bio Research at Michigan State University (Project No. MICL 02013).
References
- Allen TW, Bradley CA, Sisson AJ, Byamukama E, Chilvers MI, et al. (2017) Soybean yield loss estimates due to diseases in the United States and Ontario, Canada, from 2010 to 2014. Plant Health Progress 18(1): 19-27.
- Hartman GL, Rupe JC, Sikora EJ, Domier LL, Davis JA, et al. (2016) Compendium of soybean diseases and pests, 5th
- Rojas JA, Jacobs JL, Napieralski S, Karaj B, Bradley CA, et al. (2017) Oomycete species associated with soybean seedlings in North America-Part II: diversity and ecology in relation to environmental and edaphic factors. Phytopathology 107(3): 293-304.
- Broders K, Lipps P, Paul P, Dorrance A (2007) Characterization of Pythium associated with corn and soybean seed and seedling disease in Ohio. Plant Disease 91(6): 727-735.
- Ellis ML, McHale LK, Paul PA, Martin SK, Dorrance AE (2013) Soybean Germplasm resistant to and molecular mapping of resistance quantitative trait loci derived from the soybean accession PI 424354. Crop Science 53(3): 1008-1021.
- Schlub RL, Lockwood J (1981) Etiology and epidemiology of seedling rot of soybean by Pythium ultimum. Phytopathology, 71: 134-138.
- Zhang B, Chen W, Yang X (1998) Occurrence of Pythium species in long-term maize and soybean monoculture and maize/soybean rotation. Mycological Research 102(12): 1450-1452.
- Broders KD, Lipps PE, Ellis ML, Dorrance AE (2009) Pythium delawarii-a new species isolated from soybean in Ohio. Mycologia 101(2): 232-238.
- Ploetz R, Mitchell D, Gallaher R (1985) Population dynamics of soilborne fungi in a field multicropped to rye and soybeans under reduced tillage in Florida. Phytopathology 75(12): 1447-1451.
- Urrea K, Rupe J, Rothrock C (2013) Effect of fungicide seed treatments, cultivars, and soils on soybean stand establishment. Plant Disease 97(6): 807-812.
- Jiang Y, Haudenshield J, Hartman G (2012) Characterization of Pythium from soil samples in Illinois. Canadian Journal of Plant Pathology 34(3): 448-454.
- Zhang B, Yang X (2000) Pathogenicity of Pythium populations from corn-soybean rotation fields. Plant Disease 84(1): 94-99.
- Zitnick Anderson KK, Nelson BD (2015) Identification and Pathogenicity of Pythium on Soybean in North Dakota. Plant Disease 99(1): 31-38.
- Campa A, Vega PE, Pascual A, Ferreira JJ (2010) Genetic analysis and molecular mapping of quantitative trait loci in common bean against Pythium ultimum. Phytopathology 100(12): 1315-1320.
- Pankhurst CE, McDonald HJ, Hawke BG (1995) Influence of tillage and crop rotation on the epidemiology of Pythium infections of wheat in a red-brown earth of South Australia. Soil Biology and Biochemistry 27(8): 1065-1073.
- Bradley C (2008) Effect of fungicide seed treatments on stand establishment, seedling disease, and yield of soybean in North Dakota. Plant Disease 92(1): 120-125.
- Wise KA, Henson RA, Bradley CA (2009) Fungicide seed treatment effects on seed-borne Ascochyta rabiei in chickpea. Hort Technology 19(3): 533-537.
- Esker PD, Conley SP (2012) Probability of yield response and breaking even for soybean seed treatments. Crop Science 52(1): 351-359.
- Huang H, Erickson R (2007) Effect of seed treatment with rhizobium leguminosarum on Pythium damping-off, seedling height, root nodulation, root biomass, shoot biomass, and seed yield of pea and lentil. Journal of Phytopathology 155(1): 31-37.
- Dorrance AE, Berry SA, Lipps PE (2004) Characterization of Pythium from three Ohio fields for pathogenicity on corn and soybean and metalaxyl sensitivity. Plant Health Progress 5(1): 10.
- Rod KS, Walker DR, Bradley CA (2018) Evaluation of major ancestors of North American soybean cultivars for resistance to three Pythium Species that cause seedling blight. Plant Disease 102(1): 2241-2252.
- Klepadlo M, Balk CS, Vuong TD, Dorrance AE, Nguyen HT (2019) Molecular characterization of genomic regions for resistance to Pythium ultimum ultimum in the soybean cultivar Magellan. Theor Appl Genet 132(2): 405-417.
- Lin F, Wani SH, Collins PJ, Wen Z, Gu C, et al. (2018) Mapping Quantitative Trait Loci for Tolerance to Pythium irregulare in Soybean (Glycine max L.). G3 (Bethesda) 8(10): 3155-3161.
- Serrano M, Robertson AE (2018) The Effect of Cold Stress on Damping-Off of Soybean Caused by Pythium sylvaticum. Plant Disease 102(11): 2194-2200.
- Serrano M, McDuffee D, Robertson AE (2018) Damping-off caused by Pythium sylvaticum on soybeans subjected to periods of cold stress is reduced by seed treatments. Canadian Journal of Plant Pathology 40(4): 571-579.
- Matthiesen RL, Ahmad AA, Robertson AE (2016) Temperature affects aggressiveness and fungicide sensitivity of four Pythium that cause soybean and corn damping off in Iowa. Plant Disease 100(3): 583-591.
- Stasko AK, Wickramasinghe D, Nauth BJ, Acharya B, Ellis ML, et al. (2016) High-density mapping of resistance QTL toward Phytophthora sojae, Pythium irregulare, and Fusarium graminearum in the same soybean population. Crop Science 56(5): 2476-
- Urrea K, Rupe J, Chen P, Rothrock CS (2017) Characterization of seed rot resistance to in soybean. Crop Science 57(3): 1394-1403.
- Rosso ML, Rupe JC, Chen P, Mozzoni LA (2008) Inheritance and genetic mapping of resistance to damping-off caused by Pythium aphanidermatum in ‘Archer’ soybean. Crop Science 48(6): 2215-
- Howell C (1991) Biological control of Pythium damping-off of cotton with seed-coating preparations of Gliocladium virens. Phytopathology 81(7): 738-741.
© 2020 Dechun Wang. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






