- Submissions

Full Text
Modern Concepts & Developments in Agronomy
On the Effectivity of Gene Silencing Induction in Fall Armyworm (Spodoptera Frugiperda) Using Specific dsRNA
Niz Jose1, Pedarros Analía1, Lewi Dalia3, Hopp Esteban2 and Salvador Ricardo1*
1Institute of Microbiology and Agricultural Zoology (IMYZA-CICVyA), Instituto Nacional de Tecnología Agropecuaria (INTA), Argentina
2Institute of Agrobiotechnology and Molecular Biology (IABIMO-CICVyA)- Instituto Nacional de Tecnología Agropecuaria (INTA), Agrobiotechnology Lab (FBMC-FCEyN), Universidad de Buenos Aires,, Argentina
3Institute of Genetics (IGEAF-CICVyA), Instituto Nacional de Tecnología Agropecuaria (INTA), Argentina
*Corresponding author: Salvador, Ricardo, Institute of Microbiology and Agricultural Zoology (IMYZACICVyA), Instituto Nacional de Tecnología Agropecuaria (INTA). Nicolás Repetto and Reseros s/n, 1686 Hurlingham, Provincia de Buenos Aires, Argentina
Submission: December 21, 2019;Published: January 09, 2020

ISSN 2637-7659Volume5 Issue4
Abstract
The fall armyworm, Spodoptera frugiperda (JE Smith)) is a polyphagous pest of maize and horticultural crops in the Americas, but it recently was found in Africa as well. Main control strategies are based on the use of chemical insecticides or transgenic crops expressing Bacillus thuringiensis (Bt) toxins. However, in the last years, resistance of S. frugiperda field populations to Bt corn transgenic was detected in different Latin American countries. The available genomic information of S. frugiperda allows the development of new control technologies based on the use of RNA interference (RNAi). Three different S. frugiperda target genes were amplified and cloned to perform in vivo synthesis of specific dsRNA (ribosomal, trehalase and tubulin genes). Second to fourth larval instars were either fed, injected or soaked in dsRNA solutions and the effect on the target gene transcripts was evaluated by reverse transcriptase quantitative PCR (RT-qPCR). Only soaking second-instar larvae in dsRNA solution of ribosomal RNA gene sequence showed a statistically significant knockdown of target mRNA levels compared to control treatment. In contrast, no significant differences were registered on third and fourth larval stages. Therefore, the results showed that RNAi responses by soaking method depend of life stages, the method of inoculation and target gene sequence. Further research is needed in order to evaluate the use dsRNA as sprayable pesticide in fall armyworm strategies.
Keywords: RNA interference; Fall armyworm; Soaking method; Pest control; Maize
Introduction
The fall armyworm (FAW), Spodoptera frugiperda (Lepidoptera: Noctuidae), is a tropical insect and one of the most important pests in the Americas. Its distribution spans from Argentina to the United States [1-3]. Recently, FAW was reported in India [4] and the equatorial region of Africa [5,6]. S frugiperda larvae exhibit polyphagous behavior enabling them to attack many cultivated plant species [7,8]. Main plant hosts include maize, sorghum and important others crop such as cotton and soybean [9-11]. At present, FAW is mainly controlled by applying chemical pesticides and/or cultivating transgenic crop expressing Bacillus thuringiensis entomotoxins (Bt). In recent years, the emergence of genetic resistance of S. frugiperda populations to Bt transgenic corn was reported in Puerto Rico, Brazil and Argentina [12-14]. Therefore, the evaluation of new control strategies for FAW control has gained renewed interest. RNAi has emerged as a promising biotechnological tool for the control of pests [15,16]. RNAi is a post-transcriptional gene silencing mechanism that involves double-stranded RNA (dsRNA) directed against a target gene or its promoter region. It is a conserved cellular mechanism described in eukaryotic organisms [17].
Despite the successful examples of RNAi uses in different insects, some reports showed that dsRNA administration fails in producing knockdown of specific target genes [18]. Most of the coleopteran’s insects showed an efficient RNAi response by dsRNA feeding or injection. In contrast, RNAi is less efficient in lepidopteran insects displaying varying effects in different species [19]. Therefore, a much larger amount of dsRNA is required to trigger RNAi response in lepidopteran insects compared to coleopteran species [18]. In previous studies, RNAi strategy was successfully used in S. frugiperda to evaluate gene functions by dsRNA microinjections [20] although more recent experiments demonstrated that accumulation of dsRNA in midgut cells endosomes contributes to inefficient RNAi effect in FAW [21]. In this study, the effect of dsRNA against FAW targeted genes of S. frugiperda was evaluated by feeding, microinjection and soaking dsRNA delivery methods. The present findings suggest that dsRNA soaking could be an effective method to produce gene suppression in FAW and represents a potential strategy for the control of this insect pest.
Material and Methods
Table 1: Primer sequences for S. frugiperda gene fragments RT-PCR amplification and β-actin RT-qPCR.
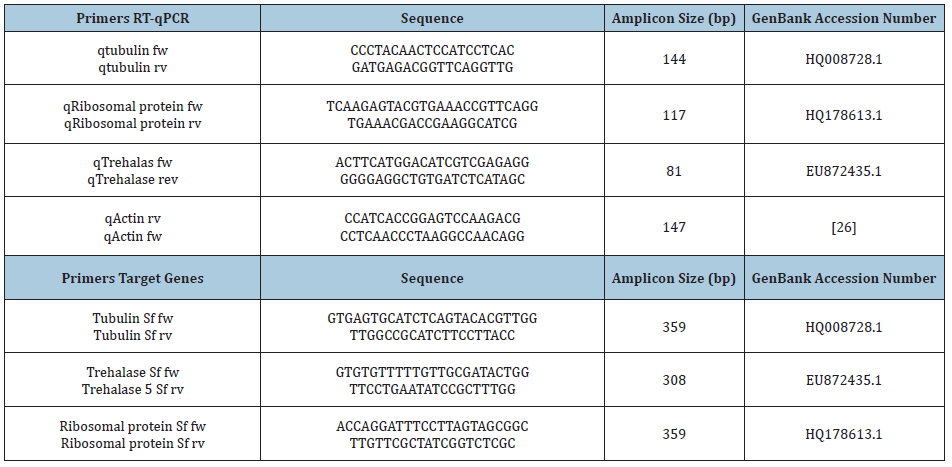
S. frugiperda larvae were reared at the Institute of Microbiology and Agricultural Zoology (IMYZA), INTA bioterium. Larvae were raised in artificial diet without formalin [22]. RNA extractions were performed on sets of 5 midguts from 3rd instar S. frugiperda larvae. Each midgut section was individually dissected and gently rinsed to eliminate the midgut diet residues. Total RNA was isolated using TRIZOL reagent kit (Invitrogen, USA) according to the manufacturer’s instructions. Extracted RNA was stored in 20μl double distilled water and its concentration was determined using a NanoDrop Spectrophotometer (NanoDrop Technologies, USA) (Table 1).
Reverse transcription PCR and gene fragments cloningTotal RNA extracted from midguts were treated with RQ1 RNase-free DNase (Promega, UK) in order to eliminate any residual DNA. Reverse transcription (RT) reactions were performed with Random primers (Invitrogen, USA) and 2μg RNA were used for complementary DNA (cDNA) synthesis with M-MLV reverse transcriptase enzyme (Promega, UK) according to the manufacturer’s guidelines. S. frugiperda genes fragments (ribosomal protein, trehalase and tubulin genes) were amplified using specific primers and PCR products ranging between 300 and 360 base pairs (bp) (Supplementary material). PCRs were performed under the following reaction conditions: 92 °C for 5 minutes (min), 35 cycles of 92 °C for 1min, 55 °C annealing temperature for 40 seconds (sec), 72 °C for 1min and 72 °C for 5min. The obtained PCR fragments were purified using Inbio Highway Kit® (Argentina) and individually cloned in pGemT vector (Promega, UK), according to the manufacturer’s guidelines. The cloned fragments were sequenced using T7 and Sp6 promoter primers to verify correct amplification. The fragments cloned in pGemT vector were released with restriction enzymes and cloned in dsRNA synthesis plasmid (pL4440) according methodology described in Gamboa et al. [23]. A plasmid pL4440- GFP with green fluorescent protein (GFP) gene fragment was used as control [23].
Bioassays
Colonies of E. coli HT115 bacteria containing pL4440 recombinant plasmids were individually grown and IPTG-induced under conditions described by Tian et al. [24]. The dsRNA expression in bacteria was verified by analyzing 1 milliliter (ml) of each culture. Each 1ml aliquot was centrifuged and resuspended in 200μl of TRIZOL reagent (InvitrogenTM) to obtain total RNA, according to the manufacturer’s instructions. The extracted RNA was dissolved in 30μl of distillated water and dsRNA expression product was confirmed by agarose gel electrophoresis (1%). After dsRNA expression verification, induced bacteria cells were collected and diluted with different volumes of ddH2O. The cell concentrations expression dsRNA of each target gene (ribosomal protein, trehalase and tubulin) were designated as 250x, 50x and 10x based on the dilution factors.
Feeding assaysIn order to evaluate the effect of dsRNA bioassays were carried out by feeding of 10x, 50x, and 250x to newly molted 2nd, 3rd and 4th instars S. frugiperda larvae, after 12h starvation. Larvae were fed using the droplet-feeding method described by Hughes and Wood [25]. After feeding confirmation, larvae were placed in individual containers with artificial diet during 8 days.
Soaking assaysNewly molted 2nd, 3rd and 4th instars S. frugiperda larvae were individually soak in dsRNA solution during 10 seconds three times a week. After soaking, each larva was placed in individual container with artificial diet during 8 days. A 40ng/ul dsRNA solution of each target gene was used in all experiments. DsRNA GFP solution was used as control.
Microinjection assaysThe direct injection of dsRNA solution into hemolymph using an insulin syringe was evaluated on newly molted 2nd, 3rd and 4th instars S. frugiperda larvae. After injection, each larva was placed in individual container with artificial diet during 8 days. A 40ng/ μl dsRNA solution of each target gene was used in all experiments. The dsRNA GFP solution was used as control.
Quantitative real time-PCRThe effect of specific dsRNA administration on the steady state transcriptional levels of target genes was evaluated by three delivery methods in 2nd, 3rd and 4th instars S. frugiperda larvae during 8 days. On the 8th day, total RNA was extracted from pools of 5 dissected midguts using Trizol reagent (Invitrogen, USA). cDNA was synthesized using MMLV reverse transcriptase (Invitrogen, USA) and random primers (Invitrogen, USA) according to the manufacturer’s instructions. The cDNAs were diluted 10 folds and used as template in qRT-PCR using the TransStart Green qPCR Super Mix (Transgen Biotech, China) and the following primers: qRib-fw 5´TCAAGAGTACGTGAAACCGTTCAGG 3´, qRib-rv 5´TGAAACGACCGAAGGCATCG3 ´; qTreh-fw 5´CACAGCCTCCCCTCCTAATTCC3´; qTreh-rv 5´ATCCCGACGTCTGCCAGATTCC3´; qTub-fw 5´CCCTACAACTCCATCCTCAC3 ´, qTub-rv 5´GATGAGACGGTTCAGGTTG3´ and actin as housekeeping: qActin-fw 5´CCTCAACCCTAAGGCCAACAGG´, qActin-rv 5´CCATCACCGGAGTCCAAGACG3´.
The PCR program used for all samples was as follows: 95 °C for 30sec, 40 cycles of 95 °C for 20sec, 55 °C for 20sec, 60 °C for 30sec followed by melting curve steps 95 °C for 15sec, 60 °C for 15sec and a final step at 95 °C for 15s. All qRT-PCR experiments included three biological replicates and three technical repetitions. The qPCRs were performed in Eppendorf Realplex2 equipment and the threshold cycle (CT) values were calculated using Eppendorf software. The CT values of ß-actin were used as reference [26].
Results and Discussion
In order to evaluate the effect of dsRNA on S. frugiperda target genes, we first amplified and cloned three DNA fragments into the plasmid pL4440 in order to synthesize them in the bacterial strain HT115 (DE3). The selected target fragments belong to genes that are involved in digestion processes (trehalase) and structural components (tubulin and ribosomal protein). Each HT115 bacterium containing the specific recombinant plasmid apparently synthesized the corresponding dsRNA in agreement with the expected molecular lengths ranging between 500 and 600bp as evidenced by agarose gel electrophoresis analysis (Figure 1). The obtained dsRNA, whose concentrations ranged between 100 and 300ng/μl, were used in feeding, soaking and microinjection bioassays. In feeding bioassays, no significant reduction in relative expression of target genes mRNA level was detected in 2nd, 3rd and 4th instars S. frugiperda larvae (data not shown).
Figure 1: Bacterially expressed dsRNA. Total RNA was extracted from IPTG induced bacteria HT115(DE3) containing the recombinant plasmids. MW: Molecular weight 1000 base pairs (bp). 1. dsRNA GFP (control). 2. pL4440 plasmid empty (control). 3. Ribosomal dsRNA. 4. Tubulin dsRNA. 5. Trehalase dsRNA. 6. Asterisks indicate the dsRNA bands.
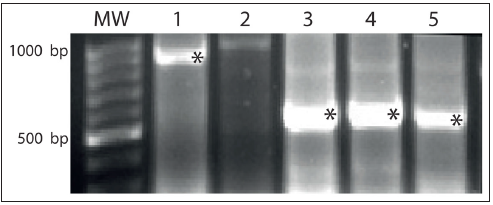
Our results are consistent with previous reports where dsRNA fed to lepidopteran larvae escapes dsRNAse digestion, enters midgut cells but it is not processed into small interference RNA [27]. Thus, gene silencing failed to occur in midgut cells. More recent evidence reveals that accumulation of dsRNA in midgut endosomes contributes to inefficient RNA interference in FAW [21]. Furthermore, similar results were obtained by using a microinjection method (data not shown). There was no significant RNAi effect in any of the instars of S. frugiperda larvae when compared to control treatments. Thus, our results do not support previous research reporting that microinjection was an effective method to produce a knock down target genes in FAW [20] although the target genes used in this study and those used in the present report are different. In contrast with Rodriguez et al. [26], recent reports showed that microinjection of dsRNA in fat bodies induce an ineffective RNAi response in FAW [21]. Therefore, further studies need to be carried out in order to clarify if the effect of dsRNA microinjection in S. frugiperda larvae is dependent of specific conditions like target gene sequence, its relative accumulation of the mRNA and other specific or general factors. Second to fourth larval instars of FAW were soaked in dsRNA solutions and the effect on the target gene transcripts was evaluated by reverse transcriptase quantitative PCR (RT-qPCR).
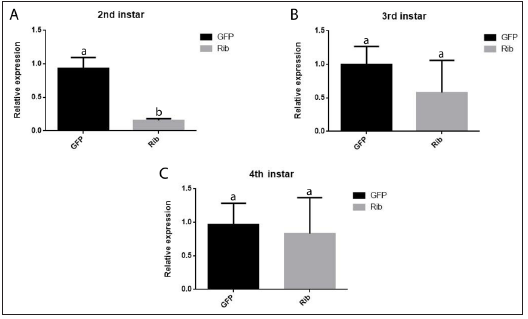
Only second-instar larvae soaked in dsRNA solution of the ribosomal RNA gene sequence showed a statistically significant knockdown of target mRNA levels when compared to the control treatment (Figure 2A). In contrast, no significant differences were registered for third and fourth larval stages (Figure 2B & 2C). Therefore, the assay results showed that RNAi responses by soaking depend on specific life stages and target gene sequence. Our experiments thus demonstrate a distinct RNA effect in FAW by dsRNA in contact with larval tegument. This result is in agreement with recent studies reporting that sprayable dsRNAs have demonstrated the feasibility and efficacy of RNAi-based gene silencing through non-transformative delivery strategies in different systems [28].
Figure 2: Relative target gene expression level in 2nd, 3rd and 4th instars S. frugiperda larvae soaked either in ribosomal (Rib) or GFP (control) dsRNAs. Bars (±SE) followed by the same letter are not statistically different at p<0.05.
Conclusion
This study demonstrated the effect of dsRNA targeted against expressed genes in S. frugiperda larvae. Here, different dsRNA delivery methods were used and RNAi effect on target genes was evaluated. In accordance with previous studies, oral and injection dsRNA administration failed to produce gene silencing on FAW. Our results indicate that a significant target gene knockdown could be achieved just only in second larval instars using a soaking method. This is the first report of such effect of gene silencing by soaking S. frugiperda larvae in dsRNA solutions. This study provides the basis for developing new FAW management methods based on RNAi. Further research is needed in order to evaluate the use dsRNA as sprayable pesticide in S. frugiperda control strategies.
Acknowledgement
We thank to Marcelo Farinón and Adrián Consiglio for insect rearing.
Conflict of Interest
The authors declare that they have no conflict of interest.
Funding
This the work was supported by the National Institute of Agricultural Technology (INTA).
References
- López EM, Hernández MJL, Rubio AP, Ochoa JM, Gutiérrez RL, et al. (1999) Biological differences between five populations of fall armyworm (Lepidoptera: Noctuidae) collected from corn in Mexico. Florida Entomologist 8(2): 254-262.
- Prowell DP, McMichael M, Silvain JF (2004) Multilocus genetic analysis of host use, introgression, and speciation in host strains of fall armyworm (Lepidoptera: Noctuidae). Annals of the Entomological Society of America 97(5): 1034-1044.
- Murúa G, Molina Ochoa J, Coviella C (2006) Population dynamics of the Fall armyworm, Spodoptera frugiperda (Lepidoptera: Noctuidae) and its parasitoids in northwestern Argentina. Florida Entomologist 89(2): 175-182.
- Ali S, Masroor Z, Masroor MD (2018) First record of the fall armyworm, Spodoptera frugiperda (JE Smith, 1797) (Lepidoptera: Noctuidae), an evil attack on paddy in Magadh, Bihar (India). International Journal of Emerging Technologies and Innovative Research 5(12): 546-549.
- Goergen G, Kumar PL, Sankung SB, Togola A, Tamo M (2016) First report of outbreaks of the fall armyworm Spodoptera frugiperda (JE Smith) (Lepidoptera, Noctuidae), a new alien invasive pest in West and Central Africa. PloS one 11(10): e0165632.
- Sisay B, Simiyu J, Mendesil E, Likhayo P, Ayalew G, et al. (2019) Fall armyworm, Spodoptera frugiperda infestations in East Africa: Assessment of damage and parasitism. Insects 10(7): pii: E195.
- Casmuz, A, Juárez ML, Socías MG, Murúa MG, Prieto S, et al. (2010) Revisión de los hospederos del gusano cogollero del maíz, Spodoptera frugiperda (Lepidoptera: Noctuidae). Revista de la Sociedad Entomológica Argentina 69(3-4): 209-231.
- Montezano DG, Specht A, Sosa GDR, Roque SVF, Sousa SJC, et al. (2018) Host Plants of Spodoptera frugiperda (Lepidoptera: Noctuidae) in the Americas. African Entomology 26(2): 286-300.
- Pitre HN, Hogg DB (1983) Development of the fall armyworm on cotton, soybean and corn [Spodoptera frugiperda]. Journal of the Georgia Entomological Society 18: 187-194.
- Bueno RC, Bueno Ade F, Moscardi F, Parra JR, Hoffmann CCB (2011) Lepidopteran larva consumption of soybean foliage: basis for developing multiple‐species economic thresholds for pest management decisions. Pest Manag Sci 67(2): 170-174.
- Hardke JT, Lorenz GM, Leonard BR (2015) Fall armyworm (Lepidoptera: Noctuidae) ecology in southeastern cotton. Journal of Integrated Pest Management 6(1).
- Storer NP, Kubiszak ME, Ed King J, Thompson GD, Santos AC (2012) Status of resistance to Bt maize in Spodoptera frugiperda: lessons from Puerto Rico. J Invertebr Pathol 110(3): 294-300.
- Monnerat R, Martins E, Macedo C, Queiroz P, Praca L, et al. (2015) Evidence of field-evolved resistance of Spodoptera frugiperda to Bt corn expressing Cry1F in Brazil that is still sensitive to modified Bt toxins. PLoS One 10(4): e0119544.
- Chandrasena DI, Signorini AM, Abratti G, Storer NP, Olaciregui ML, et al. (2018) Characterization of field‐evolved resistance to Bacillus thuringiensis‐derived Cry1F δ‐endotoxin in Spodoptera frugiperda populations from Argentina. Pest Manag Sci 74(3): 746-754.
- Price DRG, Gatehouse JA (2008) RNAi-mediated crop protection against insects. Trends in Biotechnology 26(7): 393-400.
- Zotti M, dos Santos EA, Cagliari D, Christiaens O, Taning CNT, et al. (2017) RNA interference technology in crop protection against arthropod pests, pathogens and nematodes. Pest Manag Sci 74(6): 1239-1250.
- Wilson RC, Doudna JA (2013) Molecular Mechanisms of RNA Interference. Annu Rev Biophys 42: 217-239.
- Terenius O, Papanicolaou A, Garbutt JS, Eleftherianos I, Huvenne H, et al. (2011) RNA interference in Lepidoptera: an overview of successful and unsuccessful studies and implications for experimental design. J Insect Physiol 57(2): 231-245.
- Huvenne H, Smaghe G (2010) Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: a review. J Insect Physiol 56(3): 227-235.
- Griebler M, Westerlung SA, Hoffman KH, Meyerin Vos M (2008) RNA interference with the allatoregulating neuropeptide genes from the fall armyworm Spodoptera frugiperda and its effects on the JH titer in the hemolymph. J Insect Physiol 54(6): 997-1007.
- Yoon JS, Gurusamy D, Palli SR (2017) Accumulation of dsRNA in endosomes contributes to inefficient RNA interference in the fall armyworm, Spodoptera frugiperda. Insect Biochem Mol Biol 90: 53-60.
- Greene GL, Leppla NC, Dickerson WA (1976) Velvetbean caterpillar: A rearing procedure and artificial medium. Journal of Economic Entomology 69(4): 487-488.
- Cedeño AG, Nakaya P, Niz J, Sciocco Cap A, Salvador R (2015) Double-stranded RNA synthesized in bacteria can be transferred to bee and varroa tissues. Journal of Apicultural Research 54(2): 99-100.
- Tian H, Peng H, Yao Q, Chen H, Xie Q, et al. (2009) Developmental control of a lepidopteran pest Spodoptera exigua by ingestion of bacteria expressing dsRNA of a non-midgut gene. PLoS One 4(7): e6225.
- Hughes PR, Wood HA (1981) A synchronous peroral technique for the bioassay of insect viruses. J Inv Pathol 37(2): 154-159.
- Rodriguez Cabrera L, Trujillo Bacallao D, Borras Hidalgo O, Wright DJ, Aira Pardo C (2010) RNAi-mediated knockdown of a Spodoptera frugiperda trypsin-like serine-protease gene reduces susceptibility to a Bacillus thuringiensis Cry1Ca1 protoxin. Environ Microbiol 12(11): 2894-2903.
- Shukla JN, Kalsi M, Sethi A, Narva KE, Fishilevich E, et al. (2016) Reduced stability and intracellular transport of dsRNA contribute to poor RNAi response in lepidopteran insects. RNA Biol 13(7): 656-669.
- Cagliari D, Dias NP, Galdeano DM, Dos Santos EA, Smagghe G, et al. (2019) Management of pest insects and plant diseases by non-transformative RNAi. Front Plant Sci 10: 1319.
© 2019 Salvador Ricardo. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






