- Submissions

Full Text
Modern Approaches in Drug Designing
Chronic Rhinosinusitis and the Gut-Sinus Axis: Evolving Strategies for Evidence-Based Medicine
Jad Makuch*
Department of Ecosystem Science and Policy, University of Miami, USA
*Corresponding author:Jad Makuch, Department of Ecosystem Science and Policy, University of Miami, USA
Submission: February 09, 2024;Published: March 13, 2024

ISSN: 2576-9170 Volume4 Issue3
Abstract
As a microbiome colonizer, Staphylococcus aureus (S. aureus) has many ways to travel from sinusitis and paranasal cavities down to the gut. Once in the gut and transformed into a pathogen, S. aureus has several ways to cause harm and may travel to other microbiomes in the human body by escaping immune defenses due to unrecognized movement. While a good deal is known about treating S. aureus in chronic rhinosinusitis, treatments for the gut remain to be explored fully. The number and functional structure of reservoirs in the gut where S. aureus may reside undetected, may provide important information about the timing and nature of transformations from colonizer to pathogen status. Studies of S. aureus reservoirs may address how immune defenses are reduced, gut permeability is increased, transformation from colonizer to a pathogen is triggered, or gram-negative bacteria that release bacterial lipopolysaccharides induce chronic rhinosinusitis. Creating databases for the study of S. aureus in the gut that embed data analytics and machine/deep learning can create a brighter drug development future. This rapid evolution of new approaches to correct existing deficiencies in drug development requires awareness of important therapeutic strategies and their nuances within a complex drug development environment. Some issues are highlighted.
Keywords:Microbiome; Gut-sinus axis; Staphylococcus aureus; Evidence-based medicine
Social Media Summary: Creating databases for the study of S. aureus in the gut that embed data analytics and machine/deep learning can create a brighter drug development future.
Introduction
The human microbiome includes a complex array of microorganisms and environments that play a crucial role in a human’s health. In the United States (U.S.), the prevalence of Chronic Rhinosinusitis (CRS) has been estimated to be roughly 10% of the entire U.S. population, according to large epidemiological studies [1]. CRS substantially impacts subjects’ quality of life, and the national financial burden has been estimated to exceed USD $20 billion annually [2]. CRS is defined as 12 weeks or more of inflammation of the paranasal sinuses and sinonasal mucosa. Categorized as an inflammatory disease, CRS is a heterogeneous group of related clinical syndromes characterized by chronic inflammation of the mucosa of the nasal cavities, paranasal sinuses, and often the lower respiratory tract. Managing the symptoms of CRS continues to be a medical challenge due to newly identified potential causes of disease and the identification of therapeutic targets. For example, gram negative bacteria that release bacterial Lipopolysaccharides (LPS) enter many tissues and may induce chronic rhinosinusitis [3]. Studies of S. aureus may also include LPS for the treatment of CRS. Examining sinonasal microbiota has become a key component in understanding CRS development and, conversely, prevention of CRS. This examination is an especially challenging task because CRS comprises an array of related but distinct inflammation-related outcomes, including nasal drainage, nasal obstruction, accumulation of mucus, nasal node development, and sinus pressure. The primary goals of this paper are to examine the current knowledge about the influence of the Upper Respiratory Tract (URT) microbiome focusing primarily on CRS, to summarize the nature and extent of evidence supporting a GI-respiratory (gut-sinus) axis influenced by Staphylococcus aureus (S. aureus), and to provide a perspective on next-generation evidence- based approaches for treating gut-sinus infections influenced by S. aureus.
CRS Pathogenesis
An explanation for these inflammatory responses includes the host’s immune response, categorized into innate and adaptive immune system components [4]. The innate immune system responds first to an intruder pathogen and attempts to eliminate it to maintain homeostasis and prevent pathogen invasion [5]. The innate immune system contains no memory of previous responses - its components usually include physical barriers like the skin, chemical barriers, and humoral components such as proteins. One CRS pathogenesis theory is the immune barrier hypothesis [6]. This theory proposes that a defective host mucosal barrier and the innate immune response predispose CRS patients to mucosal inflammation when colonized by bacteria that would otherwise not harm human health (i.e., commensal bacteria). The innate system also activates the adaptive immune system. The host’s adaptive immune system functions as the next level of defense if the pathogen persists due to the failure of the innate system to disarm it. The adaptive system is highly specific to a particular antigen and can provide long-lasting immunity with its components, including immunoglobulins and T-cell receptors. The adaptive system responds in various ways, but one key role is activating T-helper (Th) cells such as Th1, Th2, Th17, and Treg [7]. Suzaki et al. [7] claim that CRS with nasal polyps is Th2-dominant, unlike a healthy microbiome that promotes the proliferation of Th1 and Th2 and creates the correct Th1/Th2 balance. In addition, Th17 cells play a significant role in chronic allergic airway inflammation.
In summary, maintaining mucosal health and the absence of inflammation depend on the contribution of several Th-cell subsets expressed simultaneously. The second component of CRS pathogenesis and the immune barrier hypothesis is the role of microbes in the production of bacterial superantigens that directly stimulate a massive inflammatory response, biofilms believed to affect the persistent nature of CRS and mucosal inflammation, and dysbiosis (i.e., loss of beneficial microbial input and increase of pathogens in the microbiome). A common feature among these hypotheses is the presence of S. aureus, a potent pathogen with a high prevalence in CRS patients [8,9]. S. aureus plays an influential, multifactorial role in altering the microbiome of CRS subjects. In CRS, S. aureus produces toxins and superantigens that are highly inflammatory [10,11]. Due to their stability and high toxicity levels in humans, some of these superantigens are sometimes referred to as bioterrorism agents [10]. Virulence factors protect the bacteria from host immune surveillance and promote their survival in hostile host environments. S. aureus produces many different toxins that serve the same purpose of protecting it from being destroyed by the host’s hostile microbiome [10]. Production of these redundant toxins is beneficial in the sense that the pathogen has multiple layers of protection that enable a prolonged chronic Th2 inflammatory reaction.
Another factor is dysbiosis, which involves changes in the structure of the microbial community influenced by factors such as disease, infections, treatments, and gram-negative bacteria that release lipopolysaccharides [3]. Dysbiosis includes one or more of the following: the loss of beneficial microorganisms, an increasing number of potentially harmful microorganisms, and the loss of overall microbiota diversity [11,12]. During healthy homeostatic conditions, the microbiota comprises a diverse group of organisms known to benefit human health and host development. Host characteristics and external environmental conditions can alter the microbial community. The loss of organisms can lead to the overgrowth of commensal bacteria with potentially harmful pathobionts that cause harm/inflammation in the host. Reduced microbial diversity is another type of dysbiosis that can lead to disease creation or progression because there are fewer types and numbers of non-redundant mechanisms to destroy pathogens. The functional capabilities and architecture of the microbiota that produce appropriate intestinal immune responses have been altered and weakened. Tolerance to host tissue and commensals should be maintained while retaining the goal of eliminating harmful pathogens. This balancing act uses Tregs, a specialized subset of lymphocytes that hinder inflammation. Commensal organisms, widespread in the microbiome and causing no harm to the host, are associated with additional mechanisms that control inflammation through anti-inflammatory networks or induce modest protective inflammatory responses. Achieving healthy homeostasis involves an ongoing interactive balancing act of elements within the human microbiome.
Staphylococcus Aureus: The GI-Sinus Axis
The critical contribution of S. aureus in CRS and its role in inflammation is established. The remainder of this paper examines this opportunistic human pathogen within the framework of the GIsinus axis, acknowledging that other contributors to CRS exist such as gram-negative bacteria that release LPS [3]. S. aureus is a complex pathogen that can evade human immune defenses, migrate silently and go unrecognized inside the immune cells from the gut to other parts of the body, such as the bloodstream [13]. Finding protective niches in the human body to hide or lay dormant for significant periods of time, S. aureus represents a challenge to maintaining microbiome homeostasis. These protective characteristics make it a significant population member and a potential threat to the human microbiome. S. aureus plays a vital role in the fundamental inflammation component of CRS and has led to detailed studies of the presence and activities of S. aureus in the nasal and paranasal microbiomes. Some interesting findings arise from these studies. Since CRS is associated with clinical outcomes such as nasal drainage and mucus production, the throat can become a pathogen pathway to other microbiomes, including the gut. CRS and its link to S. aureus may cause dysbiosis in different microbiomes and lead to additional health concerns such as a ‘leaky gut.’ Upon colonization in the nasopharynx, there are complex interactions of S. aureus with host epithelial cells, mucus layer, nasal microbiota, and immune cells. Combined with other factors such as host-immune failure and inflammation, S. aureus translocate deeper into tissues, cavities, and blood vessels, including the gut and lungs [14]. Some sinonasal microbiomes provide a sanctuary, or reservoir, where S. aureus resides undetected and can delay infecting or reinfecting the host.
After ingestion or other forms of passage, including sinus drainage or translocation from the bloodstream, S. aureus may travel to the gut. See Figure 1. On the right-hand side of Figure 1 is a gut in a state of healthy homeostasis. Gut homeostasis is achieved through ‘complex crosstalk with the mucosa immune system… involving signaling pathways and gene regulatory networks’ [14]. What happens in the gut is similar to what occurs in the nasal and oral cavities. Colonization occurs, and S. aureus interacts with different types of intestinal cells, the mucus layer, gut microbiota, and immune cells. During this period of homeostasis, the healthy gut shows no breaks or breaches of the cell wall. GI barrier integrity prevents harmful molecules from invading the tissue. A different picture emerges when combined with other factors, such as host-immune failure and increases in gut permeability due to inflammation and dysbiosis discussed earlier. In this setting, S. aureus can cause gut damage (e.g., leaky gut) and a break in the cell wall. This is shown on the left-hand side of Figure 1. S. aureus now can translocate from the mucus layer deeper into other tissue, microbiomes, and blood vessels. The colonizer has transformed itself into an invading pathogen. How gut colonization of S. aureus becomes pathogenic and can travel to infect other parts of the host needs further study. Up to this point, examining the GI-sinus axis has focused on CRS, S. aureus, and how this pathogen impacted the gut and other body systems. Additional population-based evidence exists to support the GI-sinus axis. Finocchio et al. [15] concluded that the association between Gastritis/ Gastroesophageal Reflux Disease (GERD) and sinusitis was statistically significant – the prevalence of GERD increased 3.7-fold among those with sinusitis compared to control subjects without any nasal disturbance. A large Taiwanese study found that the risk of developing chronic rhinosinusitis was more than double in newly diagnosed GERD cases compared to matched controls. While other studies also were positive, the International Consensus Statement on Allergy and Rhinology: Rhinosinusitis concluded, after a review of these and other studies, that fair (not good) evidence supported the association between chronic rhinosinusitis and GERD [16].
Figure 1:S. aureus in the Gut: Colonizer to Pathogen and Invader. Figure 1 (see [14]) shows a model of the gut after the nasopharyngeal transfer of S. aureus and gut translocation. On the right-hand side of Figure 1 is a gut in a state of healthy homeostasis. The left-hand side of Figure 1 is a gut with a breached barrier. S. aureus now has the opportunity to translocate from the mucus layer deeper into other tissue, microbiomes, and blood vessels.
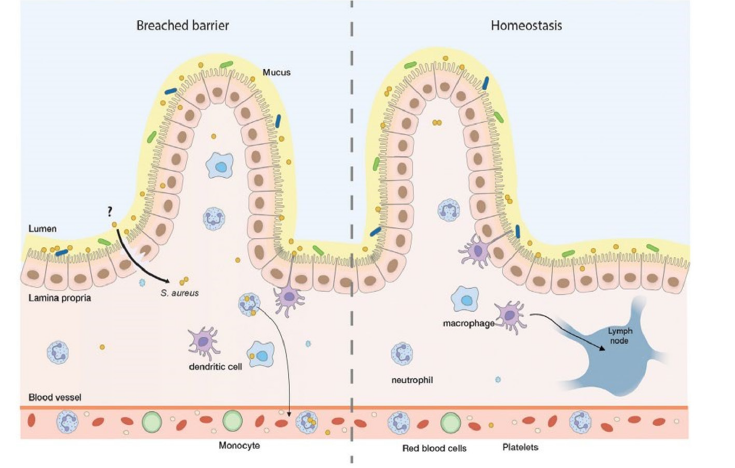
Several mechanisms were proposed to explain these observations. The first was similar to CRS in one way - namely, gastric acid exposure may exacerbate inflammation within the URT mucosa. But while the inflammation model remained applicable, there was no mention of S. aureus having any role to play. The second mechanism involved Helicobacter pylori (H. pylori), wellknown to be present in the stomach but less well-known to be found in nasal polyps and subjects with GERD and CRS. It was proposed that H. pylori causes gastritis and systemic inflammation in the nasal mucosa. Other studies have shown high levels of S. aureus in the sinus and gut mucosa of subjects with ulcerative colitis. The explanation was that patients with CRS have dysbiosis, chronic inflammation, nasal drainage, and mucus production that is swallowed. As a result, the pathogens enter the gut, where an inflammatory response occurs in response to the presence of a pathogen. Chronic inflammation and toxins in the gut eventually lead to ulcerative colitis.
Further Remarks and Next-Generation Evidence- Based Medicine
The presence of S. aureus in conjunction with CRS can lead to serious consequences affecting the human microbiome and homeostasis. Effective treatments must be identified regarding how best to treat S. aureus once it has entered the bloodstream, how to prevent CRS, how to prevent a leaky gut and epithelial dysfunction, and ways to prevent/mitigate/treat any of the harmful outcomes of S. aureus in the gut that can proceed to harm the human microbiome more broadly. A range of possible treatments for CRS depends on the subject’s precise nature of their disease (e.g., the presence/absence of nasal polyps and other comorbidities), the extent of disease control, previous treatments, and the subject’s past medical history. Current therapies target symptoms but do not provide a cure. Medical and surgical treatments include antibiotics, short-term oral corticosteroids, steroid nasal sprays, sinus irrigation, and endoscopic sinus surgery. New treatments are being studied and approved by regulators because subjects continue to have symptoms despite the availability of current treatments. For example, the U.S. Food and Drug Administration (FDA) approved the first treatment for inadequately controlled CRS with nasal polyps on June 26, 2019. The drug, dupilumab, is given by injection and provides an alternative to intranasal steroids and other treatment options. Subjects treated with dupilumab had significant reductions in their polyp size and reduced nasal congestion. This drug can reduce the need for nasal polyp surgery and oral steroids, but it is not a cure. In 2020, the European Commission approved omalizumab as an add-on therapy with Intranasal Corticosteroids (INC) for subjects with severe CRS and nasal polyps for whom INC alone did not control the disease. For a similar type of CRS subject with severe CRS and nasal polyps that could not be controlled by current therapy, the FDA approved mepolizumab (Nucala) in 2021. The issue with these newly approved therapies is that they all focus on subjects with severe CRS. New treatments are needed for subjects with milder CRS symptoms to minimize their chances of progressing to more severe forms of their disease. The development and thorough evaluation of treatments for S. Aureus intestinal colonization remains to be fully explored. There is no working S. aureus vaccine, emphasizing the need for new treatment paradigms to eliminate S. aureus from the intestine since its colonization can persist following cessation of antibiotics to treat CRS. This S. aureus persistence in the gut after nasal antibiotic treatment allows future S. aureus outbreaks to occur throughout the human microbiome.
Piewngam et al. [17] conjectured “that the mechanisms underlying nasal and intestinal colonization by S. aureus differ significantly” and that the intestine rather than the nasal domain is the primary site for initiating the harmful effects of S. aureus. Next-generation evidence-based medicine, using machine and deep learning, represents an evolutionary step [18] in sorting through such data and developing new treatment paradigms/ targeted therapeutic strategies for treating S. aureus in the gut. Figure 2 from the International Counsel for Harmonization (ICH) shows the different phases of clinical trials (i.e., the life cycle of drug development) for which a new product traditionally takes ten years to move from pre-clinical identification to approval after confirmatory Phase III trials are completed. The development cost can exceed one billion USD. Figure 2 illustrates the intersection and complexity of balancing different safety and efficacy goals during a drug’s life cycle, proceeding from pharmacokinetic/ pharmacodynamic testing in Phase 1 through various levels of therapeutic testing beginning with exploratory therapeutic studies. If these exploratory therapeutic studies are successful in certain patient populations and disease categories, large Phase III and IV post-approval studies will be conducted to confirm therapeutic activity and broader areas of therapeutic use, respectively.
Figure 2:Figure 2 from the International Counsel for Harmonization (ICH) illustrates the intersection and complexity of balancing different phases of development (Phase I through Phase IV, on the x-axis) and types of studies by objective (small Phase I pharmacology/ pharmacokinetic studies through large Phase IV studies of therapeutic use, on the y-axis). The shaded circles show the types of study most often conducted in a certain phase of drug development, and the open circles show less frequent study types conducted during that same phase of development..
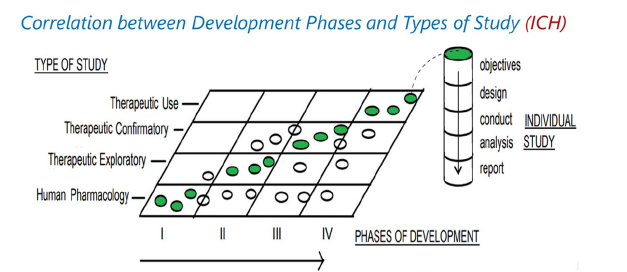
Each phase has substantively different requirements and goals. Safety is the primary focus in Phase I studies, with little interest in examining efficacy. Clinical studies proceed to Phase II, which are exploratory studies of efficacy in different patient populations while safety remains important. Phase III trials are confirmatory and focus on proving efficacy, while safety is collected to estimate benefit: risk. Conceptually distinct study designs include noninferiority and superiority designs in Phases II-IV. After approval, Phase IV studies are performed in general patient populations that are broader than in Phase III. Machine and deep learning are being proposed as new ways to increase value by shortening the developing times of successful therapeutics during Phases I-IV [18]. With the goal of designing algorithms that gather information and using this information to learn more, machine and deep learning processes can be used to create general-population Real-World Evidence (RWE) which plays an increasingly important role in healthcare monitoring of post approval safety and precision medicine [19]. The use of RWE is becoming a part of pre-approval activities, too. Machines and deep learning processes offer the possibility of creating novel algorithms and inserting fresh strategies into the existing drug development paradigm of highly structured, restrictive clinical trials [20]. The subtle changes in balancing safety vs. efficacy during a drug’s life cycle alter the nature of the studies’ design, conduct, analysis, and interpretation. The goal is to identify drugs and treatment strategies that maximize benefit: risk and meet regulatory standards. A challenge facing machine and deep learning in next-generation evidence-based medicine begins with recognizing and implementing the changing dynamics of different life cycle phases during drug development seen in Figure 2. Creating databases that embed data analytics and machine/deep learning requires an in-depth knowledge of the drug development environment. Correcting existing deficiencies in drug development requires awareness of numerous yet critically important nuances during the development cycle. The suggested use of existing study designs such as ‘N of 1’ and MOT (a combination of the master intervention trial and prospective observational trial designs) requires careful review. Some designs may be appropriate for some phases of clinical drug development and its life cycle (e.g., proofof- concept, ‘early’ vs. ‘late’ Phase II studies, pivotal studies) but not others. To achieve the hoped-for outcome of speeding clinical drug development, machine and deep learning processes will require careful development. For example, the MOT design does not provide a useful platform for designing pivotal Phase III studies because a concurrently randomized comparator group is not part of the observational study design. On the other hand, the MOT design frequently appears in post-approval Phase IV studies where the gold-standard Randomized Clinical Trial (RCT) with a concurrent comparator is no longer feasible for ethical or other reasons. Even in post-approval Phase IV studies, the MOT design can be problematic. The observational study component of the MOT design is subject to channeling, selection, and other biases do not present in a classic prospective RCT. Channeling bias occurs when clinicians prescribe one of two possible treatments based on a patient’s prognosis. In this setting, higher risk patients may be preferentially treated with one treatment, leading to a biased estimate of a treatment effect [21]. Medical researchers and regulatory agencies have embraced propensity-score analysis to eliminate or reduce this systematic bias from arising [22,23]. Propensity score analysis replaces regression analysis because it “mimics some of the particular characteristics of an RCT” [22].
Concluding Remarks
The past decades have shown the critical importance of maintaining healthy homeostasis of the human microbiome and its positive impact on human health. Health can suffer when microbiome colonizers switch and become pathogens. Immune imbalances and increases in gut permeability are factors, among many others, that can arise and lead to disease development or progression. As a microbiome colonizer, S. aureus has many ways to travel from paranasal cavities down to the gut. Once in the gut and transformed from a colonizer into a pathogen, S. aureus can cause harm and even travel to other microbiomes in the human body by escaping immune defenses. The unrecognized movement of S. aureus as a pathogen, followed by its invasion of a healthy microbiome, must be understood to develop effective new therapies that mitigate or prevent colonization or pathogen transformation. Machines and deep learning may offer useful and novel insights into next-generation evidence-based medicine. While much information is known about S. aureus in CRS, its influence on the gut requires further examination. Studies of the number and functional structure of safe harbors where S. aureus can reside, and changes in their exact location in the gut, may provide valuable information about the timing and nature of transformations from colonizer to pathogen status. Using machines and deep learning processes, new and unexpected ways may be identified to influence microbial communities and alter their functional structure to prevent/ mitigate pathogen transformation. These processes may offer novel approaches to examining how immune defenses are reduced, gut permeability is increased, or transformation triggers occur due to S. aureus colonization. Machines and deep learning may offer previously-unrecognized ways to identify new drug selection and treatment paradigms in a shortened period. In summary, a revolutionary period in evidence-based translational medicine lies ahead.
Acknowledgment
I wish to thank Professor Kathleen McCauliffe for her ongoing advice and encouragement in my exploration of the microbiome and the gut-sinus axis. I also wish to acknowledge the advice and guidance of Robert Makuch and the reviewer whose remarks led to improvements in the revised manuscript.
Author Contribution
I confirm that the research question posed, and all efforts in assembling the findings and writing the manuscript submitted were conducted by me.
Financial Support
This research received no grant from any funding agency, commercial or not-for-profit sectors.
Research Transparency and Reproducibility
I will make all data, materials, graphs, and any other information available to all readers without undue barriers to access.
References
- Hastan D, Fokkens WJ, Bachert C, Newson RB, Bislimovska J, et al. (2011) Chronic rhinosinusitis in Europe-an underestimated disease. A GA²LEN Study. Allergy 66(9): 1216-1223.
- Smith KA, Orlandi RR, Rudmik L (2015) Cost of adult chronic rhinosinusitis: A systematic review. The Laryngoscope 125(7): 1547-1556.
- Wang SB, Chen SM, Zhu KS, Zhou B, Chen L, et al. (2020) Increased lipopolysaccharides content is positively correlated with glucocorticoid receptor-beta expression in chronic rhinosinusitis with nasal polyps. Immunity Inflammation and Disease 8(4): 605-614.
- Ryu G, Kim DW (2020) Th2 inflammatory responses in the development of nasal polyps and chronic rhinosinusitis. Current Opinion in Allergy and Clinical Immunology 20(1): 1- 8.
- Riera Romo M, Pérez-Martínez D, Castillo Ferrer C (2016) Innate immunity in vertebrates: an overview. Immunology 148(2): 125-139.
- Bardy JJ, Sarovich DS, Price EP, Steinig E, Tong S, et al. (2018) Staphylococcus aureus from patients with chronic rhinosinusitis show minimal genetic association between polyp and non-polyp phenotypes. BMC Ear, Nose and Throat Disorders 18(1): 1-8.
- Suzaki H, Watanabe S, Pawankar R (2013) Rhinosinusitis and asthma-microbiome and new perspectives. Current Opinion in Allergy and Clinical Immunology 13(1): 45-49.
- Huntley KS, Raber J, Fine L, Bernstein JA (2021) Influence of the microbiome on chronic rhinosinusitis with and without polyps: an evolving discussion. Front Allergy 2: 73708.
- Boase S, Foreman A, Cleland E, Tan L, Melton-Kreft R, et al. (2013) The microbiome of chronic rhinosinusitis: culture, molecular diagnostics and biofilm detection. BMC Infectious Diseases 13(1): 210.
- Tam K, Torres VJ (2019) Staphylococcus aureus secreted toxins and extracellular enzymes. Microbiology spectrum 7(2): 10.1128.
- Seiberling KA, Conley DB, Peters AT, Grammer LC, Shuh L, et al. (2005) Superantigens and chronic rhinosinusitis: detection of staphylococcal exotoxins in nasal polyps. Laryngoscope 115(9): 1580-1585.
- Petersen C, Round JL (2014) Defining dysbiosis and its influence on host immunity and disease. Cellular Microbiology 13(7): 1024-1033.
- Tan NC, Foreman A, Jardeleza C, Douglas R, Vreugde S, et al. (2013) Intracellular staphylococcus aureus: the trojan horse of recalcitrant chronic rhinosinusitis? International Forum of Allergy & Rhinology 3(4): 261-266.
- Raineri EJM, Altulea D, van Dijl JM (2022) Staphylococcal trafficking and infection-from 'nose to gut' and back. FEMS Microbiology Reviews 46(1): 041.
- Finocchio E, Locatelli F, Sanna F, Vesentini R, Marchetti P, et al. (2021) Gastritis and gastroesophageal reflux disease are strongly associated with non-allergic nasal disorders. BMC Pulmonary Medicine 21(1): 53.
- Orlandi RR, Kingdom TT, Hwang PH, Smith TL, Alt JA, et al. (2016) International consensus statement on allergy and rhinology: rhinosinusitis. International Forum of Allergy & Rhinology 6(1): S22-S209.
- Piewngam P, Otto M (2020) Probiotics to prevent Staphylococcus aureus disease? Gut Microbes 11(1): 94-101.
- Subbiah V (2023) The next generation of evidence-based medicine. Nat Med 29: 49-58.
- Concato J, Corrigan-Curay JD (2022) Real-world evidence – where are we now? N Engl J Med 386(18): 1680-1682.
- Collins R, Bowman L, Landray M, Peto R (2020) The magic of randomization versus the myth of real-world evidence. N Engl J Med 382(7): 674-678.
- Petri H, Urquhart J (1991) Channeling bias in the interpretation of drug effects. Statistics in Medicine 10(4): 577-581.
- Austin PC (2011) An introduction to propensity score methods for reducing the effects of confounding in observational studies. Multivariate Behavioral Research 46(3): 399-424.
- Rosenbaum PR, Rubin DB (1983) The central role of the propensity score in observational studies for causal effects. Biometrika 70(1): 41-55.
© 2024 Jad Makuch. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






