- Submissions

Full Text
Modern Approaches in Drug Designing
In silico designing of Targeted Protein Degraders
Subhendu Mukherjee, Suraj T Gore, Chandrasekhar Abbineni, Murali Ramachandra and Susanta Samajdar*
Aurigene Discovery Technologies Ltd., India
*Corresponding author: Susanta Samajdar, Aurigene Discovery Technologies Ltd., Bangalore 560 100, India
Submission: December 23, 2020;Published: February 15, 2021

ISSN: 2576-9170 Volume3 Issue2
Abstract
Targeted protein degradation is projected as a radical therapeutic strategy in developing small molecule drugs. However, rational designing of such chemical entities is challenging and various CADD approaches are being explored across the globe to generate novel degraders. This brief review portrays some of the notable in silico molecular modeling techniques employed to date in degrader designing, including one of our proprietary algorithms (ALMOND) which demonstrated significant predictive ability across different target classes.
Introduction
Targeted protein degradation as a therapeutic approach has seen phenomenal
development and huge investments in recent years [1]. Proteolysis - targeting chimeras
(PROTACs) and associated molecules that induce targeted protein degradation are of
great value mainly because of potential advantages over conventional target occupancydriven
inhibitors with respect to dosing, safety, efficacy, selectivity and ability to modulate
‘undruggable’ targets [2]. These heterobifunctional small molecules harbor three chemical
features: a moiety binding to the target protein, another one binding to E3 ubiquitin ligase
and a linker for conjugating these two elements [3]. Apart from PROTACs, there are also
certain non-chimeric small molecule protein-dimerizers called Molecular Glues, which also
bind ubiquitin E3 ligases and recruit proteins for degradation, similar to PROTACs bringing
about targeted protein degradation [4].
Clinical effectiveness of molecular glues is well-known [5] and quite a few PROTAC
molecules have also recently shown adequate safety profile, therapeutic window and anticancer
activity in the clinical setting [6]. However, the know-how of chemical matter designing
is still maturing and the rational design approaches for degrader-based molecules are
currently being probed [2]. These chemical entities can be modelled along with their target
proteins as a tri-component binding system that can display cooperativity because of specific
ligand-induced molecular recognition. In the beginning, most drug design techniques in this
field relied on binary target engagement, partly due to limited structural data on ternary
complexes. However, recent co-crystal structures of several PROTACs in ternary complex
highlight the importance of protein–protein interactions and intramolecular contacts to the
mode of action of this class of compounds [4]. These discoveries have opened the door to a
new direction for structure-guided drug designing. This short perspective underscores some
of the thought-provoking and noteworthy in silico structure-guided predictive algorithms
explored so far for modeling and designing of such chemical entities.
One of the earliest known structure-guided PROTAC modeling algorithm is the PPIT
(Protein-Protein Interaction Inducing Technology) methodology of Arvinas. The technique
involves homology modeling coupled with molecular dynamics simulation that eventually
aids in de novo warhead and linker designing [7]. Pérez-Benito et al. [8] described a molecular
modeling tool that connects different pharmacophore signals via the shortest pathway along
the receptors vdW surface and then computes scores for prioritization of new bivalent
ligand designs. This tool could evaluate preferred linker lengths for different systems. Pfizer
delineated a ternary complex based restrained and exhaustive conformer sampling technique
which was used for designing BTK degraders of varying linker lengths [9]. The models
generated rationally elucidated simultaneous engagement of BTK and CRBN by PROTACs, which eventually leads to BTK ubiquitination and degradation.
Dana-Farber, Harvard Medical School and Novartis reported
application of a molecular docking tool named Rosetta [10,11] for
generating virtual models of degrader induced ternary complexes.
Using this tool, they demonstrated modest reproducibility of the
crystallographic binding conformation of a BRD4 degrader in
complex with target protein and CRBN. The model also provided a
rational direction for optimization of linker length and attachment
position (exit vector). Very recently, Bai N et al. [12] also reported
implementation of the same tool for generating models of ternary
complexes. In addition, they demonstrated that the generated models
can be translated to rational prediction of degradation potency as
well as selectivity. A 3D linker designing methodology from Oxford
Protein Informatics Group, Exscientia, Ltd and the University of
Cambridge [13] highlighted a structure-guided fragment or partial
structure linking approach. The technique considers two fragments
or fractional structures and thereafter designs a bifunctional
molecule incorporating both. The whole process is protein-contextdependent
and considers relative distance and orientation between
the partial structures. Chemical Computing Group published some
clustering and conformer sampling-based approaches for modeling
ternary complexes and demonstrated that the techniques could
reliably reproduce known crystallographic ternary complex
structures [14,15]. Another fascinating methodology proposed by
Weizmann Institute of Science employs sampling of both protein−
protein interaction as well as degrader molecule conformational
spaces. Using this methodology, they could demonstrate nearnative
prediction of crystallographic binding modes and ternary
complex conformations. The technique is known as PRosettaC
[16]. Even though some of these techniques could predict the
conformation of ternary complexes with modest to good accuracy,
there are only limited validation data available in the public domain
to comprehend the extent to which these predictive models or tools
can reliably predict secondary outcomes like ubiquitination and
protein degradation. We recently developed a computing algorithm,
ALMOND (ALgorithm for MOdeling Neosubstrate Degraders)
[17] that employs both protein-protein as well as small moleculeprotein
docking simulations along with exhaustive conformational
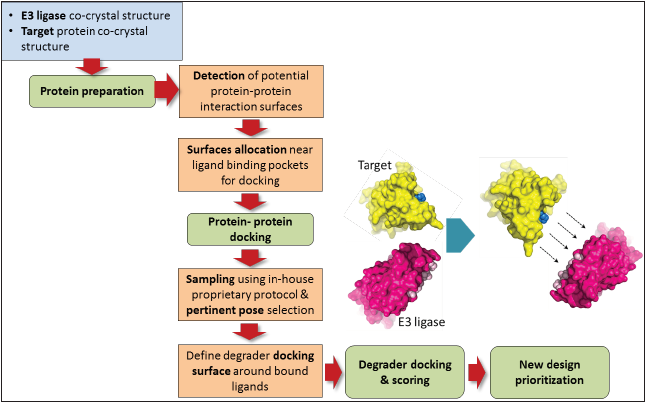
sampling and scoring. An outline of the same is depicted in Figure
1. Using this approach, we could demonstrate prediction of target
degradation potency, as well as isoform selectivity in an epigenetic
target class (SMARCA2/4) with over 80% accuracy. Good predictive
accuracy has been observed in few other target classes, as well as
BET bromodomains and kinases. Nevertheless, this technique
is currently limited to designing degraders with very short or no
linkers and further developments are ongoing to address the gaps.
Figure 1: Schematic illustration of ALMOND algorithm.
In summary, targeted protein degradation has evolved as a trailblazing technique for knocking down different classes of proteins. Despite a number of distinct advantages over traditional target inhibitors, rational designing of such class of compound remains a significant challenge as of date. The authors believe advances in in silico and structural modeling techniques will play vital roles in the near future to gain finer comprehension of the structural biology and dynamics of degrader ternary complexes and will be essential to address the current gaps in knowledge associated with such chemical matter design.
References
- Chamberlain PP, Hamann LG (2019) Development of targeted protein degradation therapeutics. Nat Chem Biol 15(10): 937-944.
- Schapira M, Calabrese MF, Bullock AN, Crews CM (2019) Targeted protein degradation: Expanding the toolbox. Nat Rev Drug Discov 18(12): 949-963.
- Gao H, Sun X, Rao Y (2020) PROTAC technology: Opportunities and xhallenges. ACS Med Chem Lett 11(3): 237-240.
- Hughes SJ, Ciulli A (2017) Molecular recognition of ternary complexes: a new dimension in the structure-guided design of chemical degraders. Essays Biochem 61(5): 505-516.
- Słabicki M, Kozicka Z, Petzold G, Li YD, Manojkumar M, et al. (2020) The CDK inhibitor CR8 acts as a molecular glue degrader that depletes cyclin K. Nature 585(7824): 293-297.
- Arvinas Press Release (2020) Arvinas releases interim clinical data further demonstrating the powerful potential of PROTAC® Protein Degraders ARV-471 and ARV-110.
- Wang J, Crew AP, Dong H, Neklesa T, Hamman B (2017) Protein-protein interaction inducing technology.
- Pérez-Benito L, Henry A, Matsoukas MT, Lopez L, Pulido D, et al. (2018) The size matters? A computational tool to design bivalent ligands. Bioinformatics 34(22): 3857-3863.
- Zorba A, Nguyen C, Xu Y, Starr J, Borzilleri K, et al. (2018) Delineating the role of cooperativity in the design of potent PROTACs for BTK. Proc Natl Acad Sci USA 115(31): E7285-E7292.
- Nowak PR, DeAngelo SL, Buckley D, He Z, Donovan KA, et al. (2018) Plasticity in binding confers selectivity in ligand-induced protein degradation. Nat Chem Biol 14(7): 706-714.
- Weitzner BD, Jeliazkov JR, Lyskov S, Marze N, Kuroda D, et al. (2017) Modeling and docking of antibody structures with Rosetta. Nat Protoc 12(2): 401-416.
- Bai N, Palani K, Karanicolas J (2020) Rationalizing PROTAC-mediated ternary complex formation using Rosetta. Bio RXiv.
- Imrie F, Bradley AR, vander Schaar M, Deane CM (2020) Deep generative models for 3D linker design. J Chem Inf Model 60(4): 1983-1995.
- Drummond ML, Williams CI (2019) In silico modeling of PROTAC-mediated ternary complexes: Validation and application. J Chem Inf Model 59(4): 1634-1644.
- Drummond ML, Henry A, Li H, Williams CI (2020) Improved accuracy for modeling PROTAC-mediated ternary complex formation and targeted protein degradation via new in silico methodologies. J Chem Inf Model 60(10): 5234-5254.
- Zaidman D, Prilusky J, London N (2020) PRosetta C: Rosetta based modeling of PROTAC mediated ternary complexes. J Chem Inf Model 60(10): 4894-4903.
- Abbineni C (2020) Targeted degradation of bromodomain-containing proteins for cancer therapy. Ubiquitin-induced targeted protein degradation session. Drug Discovery Chemistry, pp.18-20.
© 2021 Susanta Samajdar. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)













.bmp)
.jpg)
.png)
.jpg)














.png)

.png)



.png)






