- Submissions

Full Text
Modern Approaches in Drug Designing
Pyridazine Derivatives as HCV Polymerase Inhibitors: A Drug Discovery for the Treatment of Hepatitis C Virus Infection
Mohammad Asif*
Department of Pharmacy, GRD Institutes, India
*Corresponding author: Mohammad Asif, Department of Pharmacy, GRD Institutes, India
Submission: October 10, 2017;Published: July 05, 2018

ISSN: 2576-9170
Volume2 Issue2
Abstract
Activities of pyridazinones as inhibitors of the hepatitis C viral polymerase. These pyridazinones bind to the hepatitis C virus non-structural protein 5B (NS5B). Hepatitis C virus (HCV) RNA-dependent RNA polymerase, referred to as non-structural protein 5B (NS5B), is responsible for replication of the viral genome and is essential in the life cycle of the virus. Recently, active-site and allosteric NS5B inhibitors have appeared. Some 3(2H)-pyridazinones exhibited potent activity as inhibitors of genotype 1 HCV NS5B polymerase focusing on the optimization of their Drug Metabolism and Pharmacokinetics (DMPK) profiles. It has long been considered an attractive target for the development of direct acting antiviral (DAA) agents. This investigation led to the discovery of potent inhibitors with improved DMPK properties which displayed potent inhibitory activities, good stability toward human liver microsomes, and high ratios of liver to plasma concentrations. In vitro DMPK data for many of the compounds are disclosed and a crystal structure of a representative inhibitor complexed with the NS5B protein.
Keywords: Antiviral agents; NS5B polymerase; HIV-1 reverse transcriptase; Pyridazinones; Hepatitis C virus; RNA-dependent; RNA polymerase; NS5B inhibitors; DMPK profiling
Abbreviations: NS5B: Non-Structural Protein 5B; HCV: Hepatitis C Virus; DMPK: Drug Metabolism and Pharmacokinetics; DAA: Direct Acting Antiviral; RDRP: RNA-Dependent RNA Polymerase; Peg-IFN: Pegylated Interferon; RBV: Ribavirin; HLM: Human Liver Microsomes; MLM: Monkey Liver Microsomes
Introduction
Hepatitis C virus (HCV) infection is a major health problem and affects more than 200 million peoples worldwide according to the World Health Organization. An estimated 3 to 4 million people newly infected with HCV each year. The HCV, a positive single strand RNA virus of the Flaviviridae family, in the USA, HCV is expected to cause 8000–10,000 deaths yearly and most common reason is liver transplantation. The care for the treatment of HCV infection is a combination of pegylated interferon (IFN) with ribavirin. Low response rates, in particular for patients infected with genotype 1 HCV, along with significant side-effects of this therapy result in an urgent medical need for improved HCV treatments. The HCV NS5B enzyme, a virally encoded RNA-dependent RNA polymerase (RdRp), is viewed as an attractive target for the development of such direct antiviral agents due to its central role in the replication of the viral genome.
Accordingly, research focused on developing small molecule, non-nucleoside NS5B inhibitors for use in patients infected with HCV genotype 1. Long-term infection can lead to chronic liver disease, such as liver cirrhosis or hepato-cellular carcinoma and a leading cause of hepatic disease. The standard of care therapy employed pegylated interferon (peg-IFN) often in combination with the nucleoside analog ribavirin (RBV). The standard of care by support of the two HCV protease inhibitors Incivek and Victrelis remains unclear as both drugs require response-guided therapy regimens that can shorten the duration of IFN therapy in infected persons. The serious side effects and limited efficacy underline the urgent need for improved therapeutic drugs. In addition, there is no established vaccine for HCV. As a result, there is an urgent need for safe and effective therapeutic drugs that fight HCV infection and that have high genetic barrier to resistance [1-5].
The drawbacks of current HCV therapy necessitate the development of more effective anti-HCV agents, especially for patients infected with genotype 1 HCV, because response rates to this therapy are sub-optimal and are mainly low in patients infected with genotype 1 HCV. The HCV NS5B protein is a virally encoded RdRp, the activity of which is critical for the replication of the virus. Inhibitors of this enzyme therefore have potential to become novel direct antiviral agents for the valuable treatment of HCV infection. Several nucleoside and non-nucleoside NS5B inhibitors are in clinical trials. Most non-nucleoside NS5B inhibitors are bind to one of three allosteric pockets of the protein that are distinct from the active site. The computational analysis of these pockets indicated that a binding site at the palm domain was highly conserved across various genotype 1 HCV sequences About 70% of infected people will develop chronic histological changes in the liver (chronic hepatitis) with a high risk of advancing to cirrhosis or hepatocellular carcinoma. The care for HCV is a combination therapy of pegylated interferon and ribavirin. Unfortunately, this treatment provides benefit for less than 50% of genotype 1 infected patients and is associated with adverse events.
Therefore, there is an urgent need for the development of more effective HCV therapies especially for genotype 1 patients. The non-nucleoside small molecule inhibitors of genotype 1 HCV NS5B polymerase which may binds at this location and are developed small molecule non nucleoside inhibitors of HCV NS5B, a RdRp [6- 8]. The treatment with peg-IFN/RBV is often associated with major side effects including flu-like symptoms, depression, and anemia. The relatively low response rates and the significant side effect burden of current HCV therapies necessitate the discovery of more effective anti-HCV agents, mainly for treatment of patients infected with genotype 1 HCV. The HCV RdRp, NS5B is a smart target for the development of novel HCV treatments. The activity of this virally encoded enzyme is crucial for HCV replication and many nucleoside and non-nucleoside NS5B inhibitors have been described. Several molecules revealed antiviral activity in HCV infected patients. Most of the non-nucleoside NS5B inhibitors bind to one of three locations on the enzyme’s surface that are distinct from the active site. The NS5B inhibitor development efforts have focused on compounds which bind to the ‘palm’ site of the NS5B protein [9-15].
Pyridazinone Derivatives as Hcv Polymerase Inhibitors: An Antiviral Agent
Numerous classes of inhibitors are known to bind at the NS5B palm site. Of these, compound 1, containing the 1,1-dioxobenzo [1,2,4]-thiadiazine moiety, exhibits potent NS5B inhibition. The compound 1 bound to the NS5B polymerase, this class of compounds has a reasonably planar conformation with a minor rotation between the quinolinedione and benzothiadiazine rings directed by intramolecular H-bonding. The structural features are highly favorable for binding to the NS5B palm site. We imagined that removing the fused benzene ring from the quinolinedione core could afford a molecule with increased flexibility and reduced lipophilicity to improve overall physicochemical properties. Therefore, replacing the quinolinedione core of compound 1 with a 5-hydroxy-3(2H)-pyridazinone, leading to structures of type 2. The modest NS5B inhibition activity displayed by an early lead compound 3 [16].

figure 1: Compound 3 bound in the NS5B palm site. Hydrogen bonds are symbolized as dashed lines, and the residues which make up the enzyme binding sub sites are illustrated.
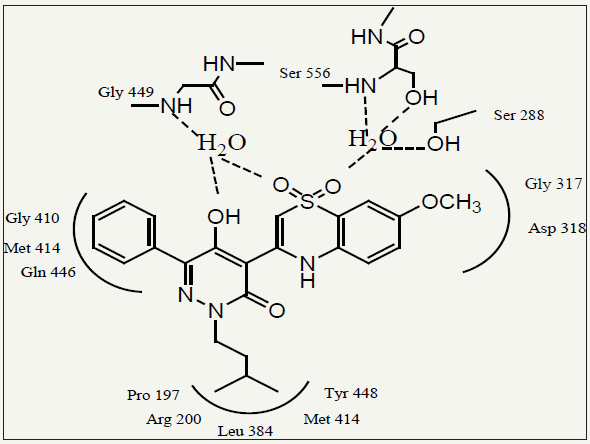
The compound 3 bound to the NS5B protein was determined in order to facilitate our subsequent lead optimization process. The structure revealed several tightly bound water molecules surrounding compound 3. Two of these water molecules mediated important H-bonds between the inhibitor and the protein. One water molecule bridged a sulfonamide O-atom to the NS5B residues Ser556 and Ser288, while the other formed H-bonds between the pyridazinone enolic OH and amino acid residue Gly449. These H-bond networks are important contributing factor for NS5B binding affinity. The key feature of the structure was a deep hydrophobic pocket in the enzyme that was occupied by the R2 isoamyl group. The R1 phenyl ring filled a fairly shallow cavity in proximity to the Met 414 side chain by rotating up to 45° out of plane relative to the pyridazinone ring. The geometry of the R1 phenyl group recommended that a smaller ring, such as a thiophene moiety, might be better accommodated in this region. The methoxy R3 substituent of 3 was deemed to be sub-optimal due to a lack of appropriate space filling. The oxygen atom at 70-position could be further elaborated to increase the NS5B binding affinity by filling up more space in the R3 region of the binding pocket (Figure 1) [16].
The lead optimization efforts by replacing the R1 phenyl ring present in compound 3 with a 2-thiophene moiety while keeping the R2 and R3 substituents unchanged (4a). This modification afforded a 9-fold improvement in NS5B inhibition activity as compared with the lead compound 3. This result is consistent with the X-ray structure analysis of compound 3 complexed with the NS5B protein. We then kept 2-thiophene as R1 and isoamyl as R2 constant but varied R3 to study the SAR at this position. The inhibitory activities against genotype 1b in both enzyme and cell culture replicon assays were measured. In addition, the stability of the compounds toward Human Liver Microsomes (HLM) was determined. Removal of the methoxy R3 substituent present in 4a led to a 3-fold increase in NS5B inhibition activity (4b), and its replacement with a hydroxyl moiety (4b) significantly improved enzyme inhibition properties. Importantly, extension of the R3 substituent along with the addition of a polar acetamide group at its terminus afforded a compound (4d) with substantially improved enzyme inhibition activity. This molecule also exhibited good antiviral potency in the replicon cell culture assay that was clearly distinct from cytotoxicity.
The significant increase in potency (>50-fold) for 4d compared with 4a can likely be attributed to the formation of additional hydrogen bonds between the acetamide moiety and the NS5B protein. Such interactions were observed in the crystal structure of 4d complexed with the NS5B protein. They involved the terminal NH of the acetamide moiety and amino acid residues Asp318 and Asp225 (via water molecules) as well as the acetamide O-atom and residues Asn291 and Asp225 (via another water molecule) of the NS5B protein. In addition, the increased length of the OCH2CONH2 (required for filling the space in the R3 region of the binding pocket) may also contribute to the increased affinity. Analysis of the 8d-NS5B structure suggested that small alkyl groups could be appended to one face of the R3 methylene moiety present in 4d without negatively impacting NS5B interactions. Accordingly, we introduced a single methyl group at this location with the expectation that this modification might reduce the polarity of the R3 moiety and increase cell permeability. The resulting compound (4e) exhibited equipotent NS5B inhibition in enzyme assays compared with the non-methylated analog 4d and displayed comparable cellular activity.
The X-ray crystal structure of 4e complexed with the NS5B protein in the R3 region clearly showed that the R3 conformation and interactions with the NS5B protein were very similar to that of 4d. In addition, as anticipated from the 4d-NS5B co-crystal structure, substitution with a germinal dimethyl moiety or larger alkyl groups (4f & 4g) resulted in a significant loss of NS5B inhibition activity. Analysis of the 8d-NS5B co-crystal structure also suggested that the R3 acetamide moiety would tolerate the addition of small appendages to its terminus. The corresponding N-methylated compound 4h and the hydroxamic acid derivative 4i which both displayed comparable NS5B inhibition activities to that exhibited by 4d. Interestingly, alkylation of the polar NH2 group of 4d with the expectation of possibly enhancing cell permeability did not improve the compound’s antiviral properties. The inclusion of a larger cyclic moiety at the R3 terminus (4j) diminished the enzyme inhibition activity by 5-fold compared with 4d, which was consistent with our prediction based on the 8d-NS5B co-crystal structure. Furthermore, replacement of the terminal amide moiety with an ester significantly reduced NS5B inhibitory potency (compare 4k with 4e) likely due to the loss of the hydrogen bonds formed between the 4e R3 terminal NH moiety and the NS5B protein. In addition, the larger R3 ethyl ester group may be too voluminous to fit well in the binding pocket. However, the inhibition activity could be partially restored by incorporation of the corresponding carboxylic acid moiety into the terminal R3 group (as in compound 4l). This result may be due to the reduction in size of the R3 group and the possible formation of hydrogen bonds between the OH group of the carboxylic acid and the surrounding NS5B residues [16].

(Table 1) Other modifications of the R3 substituents, that are either C- or N-linked to the benzothiadiazine ring (4n–4y). None of the variations including ketones, amides, heteroaromatics, and cyano moieties led to any improvement in NS5B inhibitory potency as compared with compounds 4d or 4e. The difference in activity of 4d, 4o, and 4n could be explained by the difference in the preferred conformation of the R3 groups. To allow for the favorable hydrogen bonding interactions, the R3 group needs to be close to co-planar to the benzothiadiazine ring. In 4d and 4o this is the preferred conformation. However, in 8n the lowest energy conformation will have the second methylene group rotated out of the plane of the aromatic system. In anticipation of assessing the in vivo PK properties of potent pyridazinone-containing NS5B inhibitors, we also examined the stability of these compounds toward human liver microsomes (HLM). The measured HLM half-lives for a majority of the compounds (4a–h, 4k, 4m, 4p–r, 4t, 4v–y) were relatively short (<30 min).
Table 1: Compounds 4a-4y.
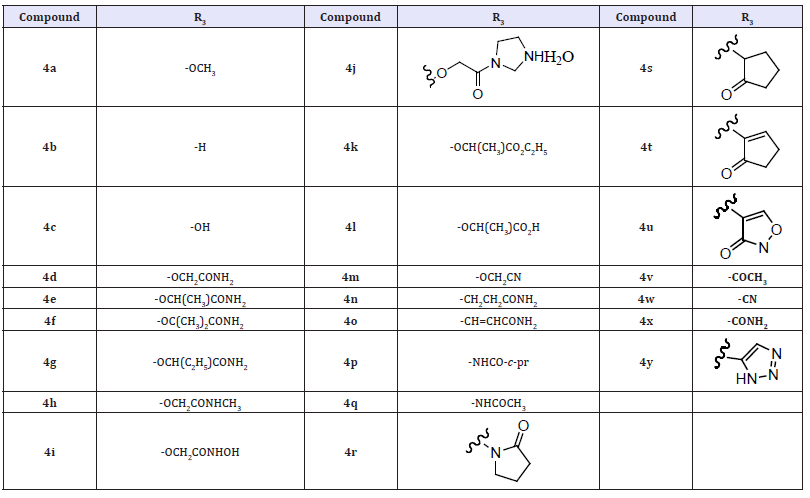
However, compounds 4j and 4s bearing either a polar cyclic acetamide moiety via an O linkage or a cyclic ketone group via a C-linkage exhibited moderate half-lives. The most stable compounds (4i, 4l, and 4n) possessed a hydroxamic acid or a carboxylic acid or a C-linked acetamide moiety and exhibited long half-lives (>60 min). These results implied that the very hydrophilic pyridazinone compounds might have poor/nonproductive interactions with CYPs and suggested that R3 substitution could impact the HML stability of the molecules under study [16]. The 4-(10, 10-dioxo- 10,40-dihydro-10. k6-benzo [10,20,40] thiadiazin-30-yl)-5- hydroxy-2H-pyridazin-3-ones as potent inhibitors of genotype 1b HCV NS5B polymerase. The new class of pyridazinones was developed. In addition, optimization of the R3 substituents in this series resulted in NS5B inhibitors with sub-micromolar potencies in both biochemical and cell culture replicon assays. Furthermore, the R3 substitution also affected the stability of the molecules toward HML [16].
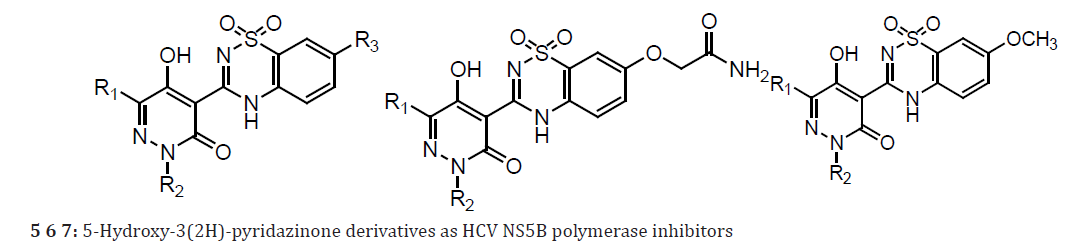
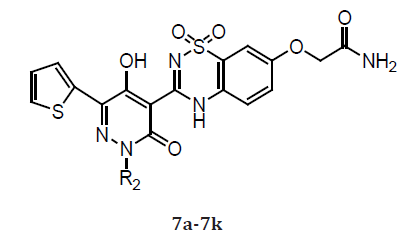
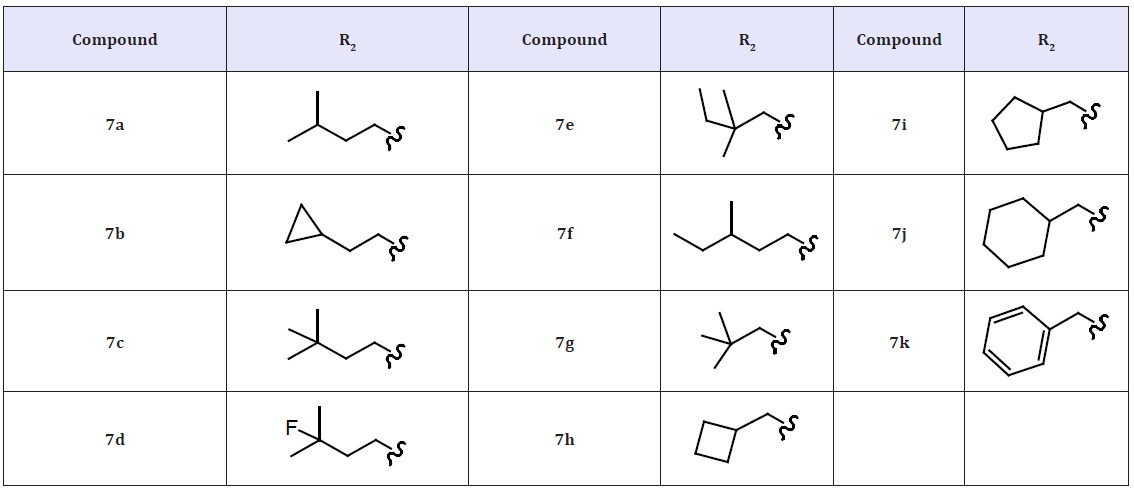
A series of 5-hydroxy-3(2H)-pyridazinones (5 & 6) using a structure-based design approach. The SAR linked with variation of the R3 substituents and identified the –OCH2CONH2 moiety as a suitable fragment for obtaining potent NS5B inhibition properties. The biological properties associated with the systematic alteration of the 2- and 6-pyridazinone substituents in a series of molecules (6) which contain this optimal R3 substituent. Pyridazinone benzothiadiazinedioxide derivatives (7) bearing a methoxy group at the 7’-position. 3(2H)-pyridazinone ester intermediates, condensation 2-amino-5-methoxy-benzenesulfonamide. The inhibitory activities of these compounds in genotype 1a and 1b NS5B enzyme assays as well as in a 1b replicon assay. Compound 7a exhibited good inhibitory potency against the genotype 1b enzyme. However, it was 5-fold less potent in the genotype 1a enzymatic assay. Compounds bearing R2 groups that were comparable in linear length to the isoamyl moiety present in 7a exhibited slightly improved activity in the biochemical assay (7b and 7c). However, they were notably less potent in the genotype 1b replicon assay compared to compound 7a. Adding an F-atom to the terminus of the isoamyl group (7d) or introducing a 2,2-dimethylbutyl group (7e) led to a significant loss in activity in the biochemical assay when compared with 7a. Interestingly, compound 7e exhibited equipotent antiviral properties in the replicon assay as compared to 7a. In addition, introduction of R2 alkyl groups that were longer or shorter than the isoamyl moiety led to compounds with NS5B inhibition activities comparable to 7a against the genotype 1b enzyme (7f and 7g).
However, these molecules were less potent than 7a in both 1b replicon and 1a enzyme assays. Furthermore, compounds bearing a methylene spacer between the pyridazinone core and a cyclic moiety displayed decreasing NS5B inhibition activities with increasing ring size (7h–7j). When compared to 7j, compound 7k having a benzyl R2 substituent displayed weaker antiviral potency in the 1b replicon assay although the inhibitory activity in the enzyme assay was improved. The different EC50/IC50 ratios observed likely resulted from the differences in cell permeability and protein binding. This series of 5-hydroxy-3(2H)-pyridazinone derivatives under study was 3- to 14-fold less potent against genotype 1a NS5B relative to the genotype 1b enzyme. We believe that this potency shift was likely due to the amino acid residue difference of Y415F (phenylalanine in genotype 1a vs tyrosine in genotype 1b) located near the R2 sub-pocket at the palm binding site [17].
The above efforts led to the identification of the R2 substituents present in compounds 2a–2c and 2h as optimal moieties for obtaining potent NS5B inhibitory activity. We then explored combinations of these R2 fragments with various R1 substituents in an effort to further improve potency. The impact of R1 substitution on the inhibitory activity against genotype 1a and 1b enzymes as well as the 1b replicon, the 3-thiophene analog (7l) was 9-fold more potent in the 1b replicon assay than the corresponding 2-thiophenecontaining compound (7b). However, introducing a methyl group at the 5-position of the 2-thiophene ring resulted in a significant loss of activities in both enzyme (30-fold) and replicon assays (4-fold) (compare 7m with 7b). This trend was observed with several other inhibitors (7n and 7o) bearing different R2 substituents (compare with 7c and 7h, respectively). The co-crystal structure of 7m bound to NS5B12 revealed that this methyl group was unable to access the small sub-pocket in the R1 region as originally intended. Instead, the methyl fragment rested on the NS5B surface and generated unfavorable interactions with nearby polar protein residues. These interactions were the likely cause of the observed decrease in the inhibition activity of 7m. A similar reduction in potency was observed by introducing a close proximity with polar protein atoms and this may explain the reduction in potency observed for 7p [17].
[Table 2]
Table 2: Compounds 7a-7k.
(Table 3) In addition, replacement of the 2-thiophene ring with a 5-thiazole moiety led to very potent compounds (7q and 7r) possessing activities comparable to 7a and 7b in both enzyme (1b and 1a) and replicon assays. This result was consistent with the analysis of the co-crystal structure of 7q bound to the NS5B protein. The pyridazinone and benzothiadiazine rings of 7q adopted a nearly co-planar geometry which was highly favorable for binding to the NS5B palm site. The 5-thiazole ring, rotated approximately 16° out of plane relative to the pyridazinone ring, was able to effectively access the sub-pocket in the R1 region. This rotation is similar to the rotation of the 2-thiophene moiety in 2a-NS5B Xray co-crystal structure. These rotations were smaller than the rotation of a phenyl R1 group (R2 = isoamyl, R3 = OMe). Due to its larger size, a phenyl ring needs to rotate more to avoid a close interaction with Gly410 and Asn411. This could explain some of the differences between the IC50 values of 7q or 7a and the phenyl substituted analogs. Additionally, a number of direct or watermediated H-bonds were formed between the sulfonamide O-atoms and residues Ser556, Ser288, and Gly449 as well as between the acetamide R3 moiety and residues Asn291 and Asp318.
Table 3: Compounds 7l-7y.
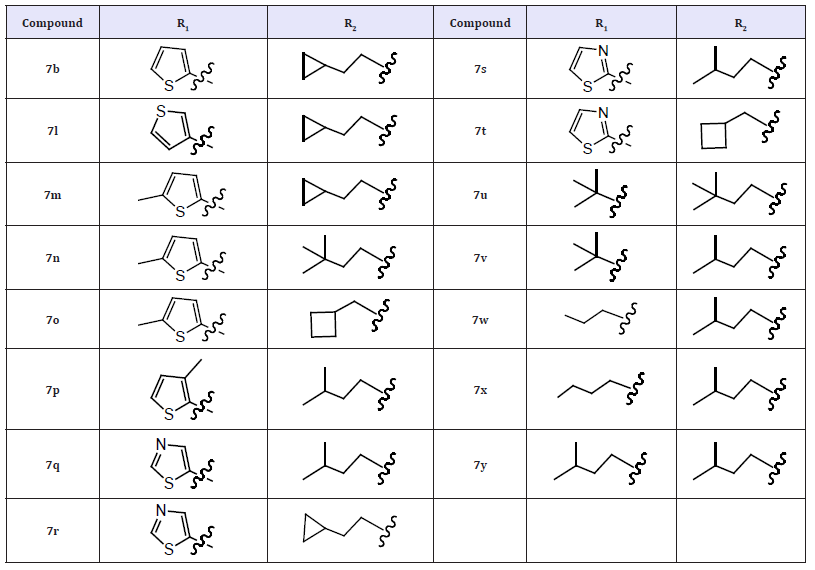
These favorable structure features, which were almost identical to those observed in the X-ray co-crystal structure of 7a-NS5B, may explain the high potency of compound 7q. (Figure 2) summarizes the observed interactions in the co-crystal structure of compound 7q bound to the NS5B protein. Surprisingly, the replacement of the 2-thiophene ring with a 2-thiazole moiety (7s and 7t) caused a significant loss in inhibitory potency in both enzyme and replicon assays when compared with 7a and 7h, respectively. The reasons for this loss in biological activity are not yet well understood. Furthermore, introduction of alkyl R1 substituents into the inhibitor design (7u–7y) did not afford any improvements in NS5B inhibitory potency as compared with compound 7a. In order to better predict the in vivo metabolic stability of this new class of pyridazinone derivatives, the effect of R1 and R2 substituents on stability toward human liver microsomes (HLM) was evaluated. The measured HLM half-life for compound 7a was relatively short (23 min).
figure 2: Diagram of compound 7q bound in the NS5B palm site. Hydrogen bonds are symbolized as dashed lines, and the residues which make up the enzyme binding subsites.
We reasoned that, in addition to the un substituted thiophene ring present in 7a, the secondary C-atom of the terminal isoamyl group might also be a potential site for cytochrome P450 (CYP) catalyzed metabolism. Accordingly, an F-atom was introduced at the secondary C-atom of the isoamyl fragment and this resulted in a more stable compound (7d). However, the addition of a methyl group at this position did not improve the stability of the resulting molecule (7c). Interestingly, inclusion of a more branched R2 group of the same linear length as the isoamyl moiety or a large cyclohexyl R2 fragment resulted in very stable compounds (7e and 7j, respectively; t1/2 > 60 min). In contrast, compound 7k, bearing a benzyl R2 moiety, had a short half-life (23 min) possibly due to CYPmediated modification of the benzene ring (compare 7k with 7j). The measured HLM half-lives for the rest of compounds (7b, 7f–7i) were relatively short. Alkyl R1 substituents led to the least stable compounds (7u, 7x) with very short half-lives (610 min). Similar to compound 7b bearing a 2-thiophene moiety, the 3-thiophene group also led to a compound (7l) with a short half-life (16 min).
As the 5-position of the 2-thiophene ring is a potential site for oxidation catalyzed by CYPs, introduce a methyl group at this position to block such metabolism. This change resulted in a significant improvement in the stability of the resulting compounds 7m, 7n, and 7o when compared with 7b, 7c, and 7h, respectively. Both 2-thiazole and 5-thiazole moieties led to very stable compounds (7q–7t) with t1/2 P 60 min. The above results indicated that both R1 and R2 substituents significantly affected the metabolic stability of the resulting molecules. The 4-(10,10-dioxo-10,40- dihydro-10k6-benzo [10,20,40] thiadiazin-30-yl)-5-hydroxy- 2H-pyrida zin-3-ones (7) as potent inhibitors of genotype 1 HCV RNA-dependent RNA polymerase (NS5B). The optimization of the 2- and 6-substituents of the Pyridazinone derivatives resulted in significantly improved potencies in both biochemical (1b and 1a) and replicon (1b) assays [17].
Inadequate response rates, in particular for patients infected with genotype 1 HCV, along with significant side-effects of current HCV therapy result in an urgent medical need for improved treatments. As part of our efforts to develop small molecule, nonnucleoside inhibitors of the HCV NS5B enzyme, a virally encoded RNA-dependent RNA polymerase (RdRp) essential for HCV replication, the previously described the discovery and structurebased optimization of a series of 5-hydroxy-3(2H)-pyridazinone derivatives which bind to the NS5B ‘palm’ site (8). In the course of this work, we identified compound 9 bearing an acetamide R3 moiety as one of the most potent NS5B inhibitors and antiviral agents [18].
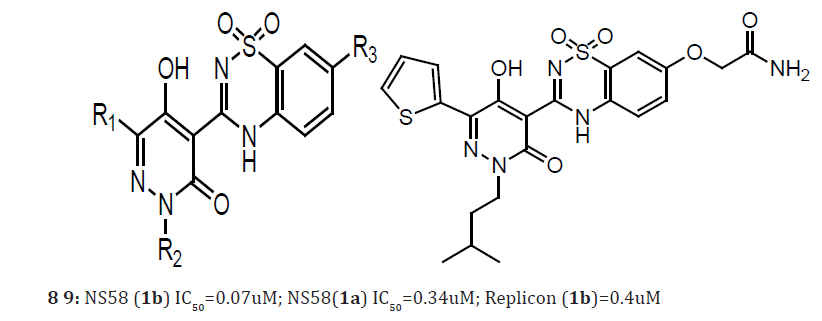
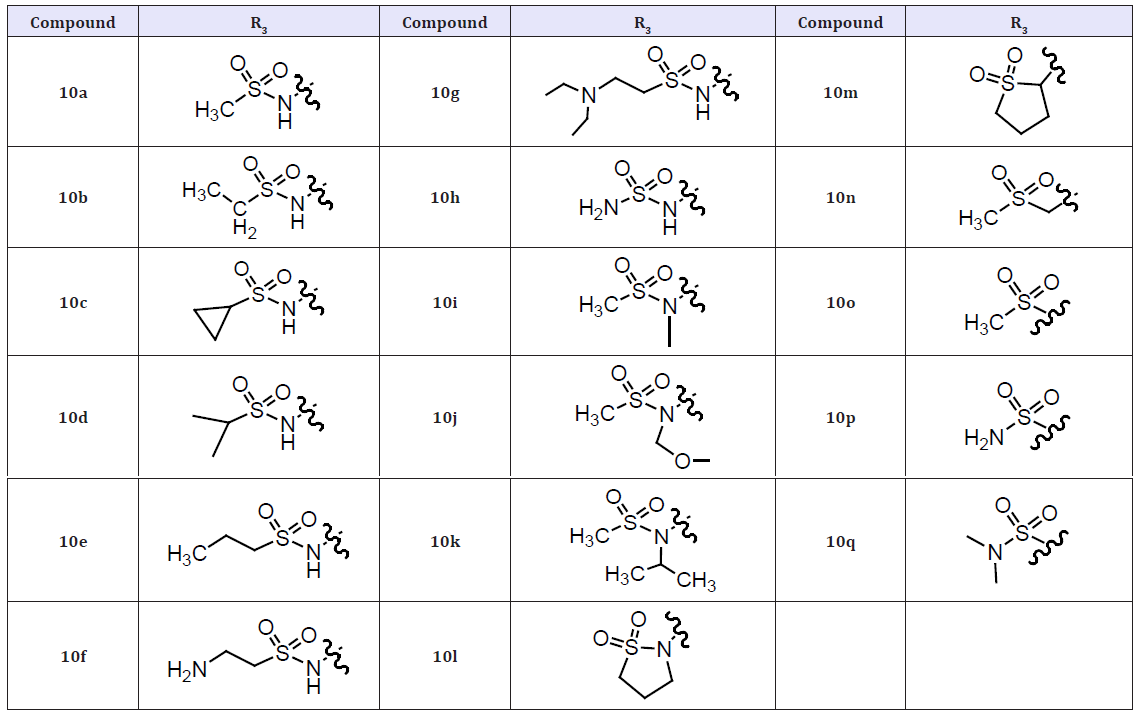
Here, we report the optimization and preliminary pharmacokinetic profiling of pyridazinone-containing NS5B inhibitors. Since during our previous studies we observed that R3 substitution had the most significant impact on inhibitor potency, we chose to continue making modifications at this position. The –OCH2CONH2 moiety present in compound 9 was replaced by a -NHSO2Me fragment, the resulting compound (10a) displayed improved NS5B inhibitory potency and antiviral activity. This significant increase in potency could be explained with the aid of the co-crystal structure of 10a bound to the NS5B protein. The -NHSO2Me R3 moiety of 10a formed H-bonds with NS5B residues Asn291 and Asp 318. Similar H-bonding interactions were also observed in the 2-NS5B co-crystal structure between the acetamide moiety of compound 9 and NS5B. Importantly, the -NHSO2Me R3 moiety of 10a formed a third H-bond network with the residue Ser288 via a water molecule that was absent in the 2-NS5B cocrystal structure.
This additional H-bond network, which complements a similar interaction from the -SO2- moiety in the benzothiadiazine ring of 10a, is likely responsible for the potency differences observed between compounds 9 and 10a. Although the above co-crystal structure indicated that the methyl substituent of the -NHSO2Me R3 moiety present in 10a was oriented away from the NS5B surface, increasing the size of this fragment was generally detrimental to anti-NS5B and antiviral properties (10b-10g). However, this methyl group could be replaced with an –NH2 moiety without sacrificing antiviral potencies (10h). Other, often drastic, losses in NS5B inhibition activity and antiviral properties were noted for compounds in which the NH of the R3 sulfonamide moiety was alkylated (10i-10l) or removed (10m-10q). Such unfavorable property alterations are likely due to the loss of the H-bond between the R3 sulfonamide NH and the NS5B Asp318 residue. The results described above confirmed the large impact that R3 variation had on the anti-NS5B and antiviral properties of this series of inhibitors and identified the –NHSO2Me and –NHSO2NH2 fragments as optimal R3 moieties [18].

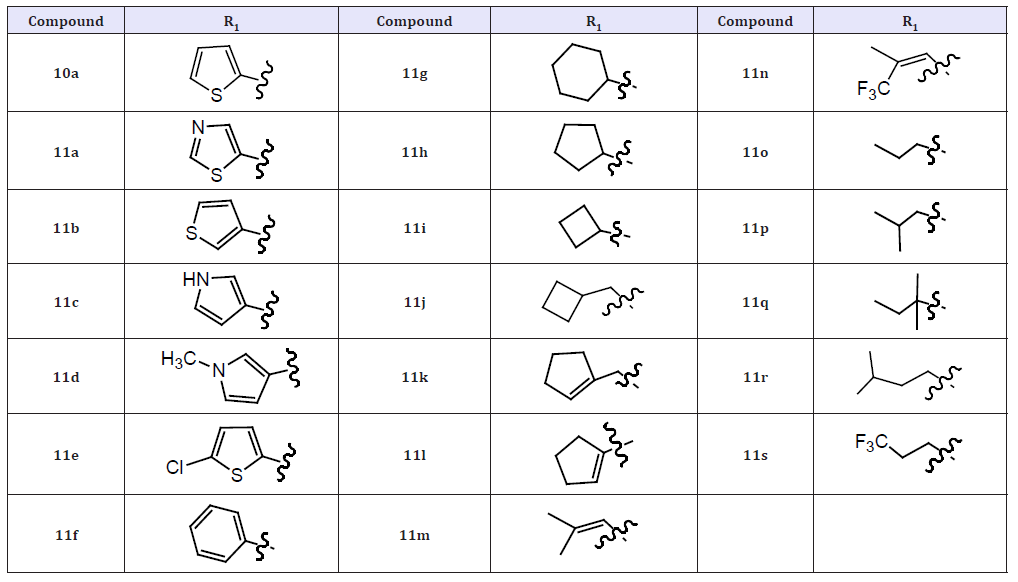
(Table 4) The systematic variation of the R1 and R2 substituents attached to the pyridazinone ring of the inhibitors. Due to its relative ease of synthesis, we employed the –NHSO2Me moiety in lieu of the -NHSO2NH2 fragment as the preferred R3 substituent. The 2-thiophene R1 group present in 10a was replaced by other unsubstituted five membered heterocycles identified during our previous studies, the resulting compounds (11a-11c) retained good NS5B inhibition activity and antiviral properties in cell culture. As we also previously observed,6 appending substituents to such heterocycles were detrimental to the associated biological potencies (11d-11e) as was the incorporation of a six-membered aromatic ring at this location (11f). Introduction of alkyl and alkenyl groups (both cyclic and acyclic) at the R1 position of the inhibitor design (11g-11s) generally afforded compounds with significantly poorer anti-NS5B and/or antiviral properties relative to 3a. The most notable exceptions were compounds 11l and 11m, which exhibited replicon potencies that were only about fourfold weaker than that displayed by 10a. While the exact reasons for these SAR are not all known, the results described above confirmed that unsubstituted five-membered heterocycles represent ideal R1 substituents that confer the most potent anti-NS5B and antiviral properties to this series of NS5B inhibitors [18].
Table 4: Compounds 10a-10q.
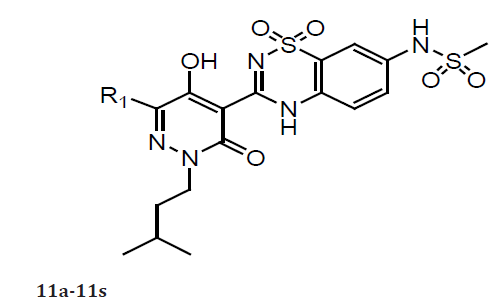
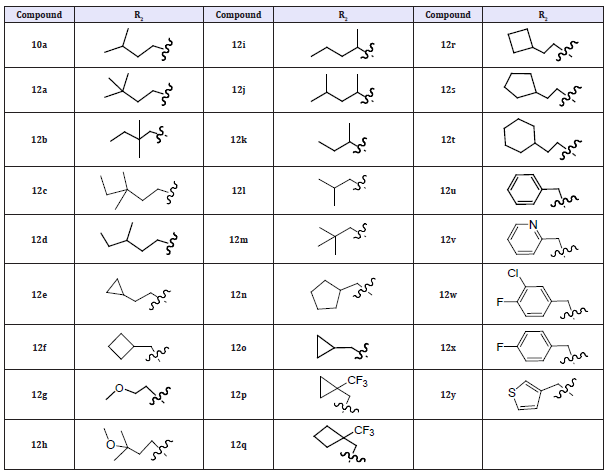
(Table 5) As previously observed, a R2 alkyl fragment in linear 4–5 carbon chain length led to both good anti-NS5B and antiviral activities (12a–12f) comparable to that of 10a. However, more polar R2 groups with a methoxy moiety at the terminus led to less potent compounds (12g and 12h) (compared with 12c). Introducing branching at the C-atom of the R2 fragment adjacent to the pyridazinone N-2 atom was highly detrimental to both anti- NS5B and antiviral properties (compounds 12i–12k). Consistent with what we observed previously,6 shorter R2 groups such as isobutyl and 2,2-dimethylpropyl (12l–12m) resulted in significant potency loss in 1b replicon assays as compared with 10a. In general, the saturated cyclic alkyl methyl/ethyl R2 groups (12n–12t) led to less potent inhibitors as compared with 10a with the exception of 12p. Interestingly, analogs containing a benzyl moiety (with or without substitution) or a 2-pyridyl methylene R2 group (12u–12x) all displayed poor antiviral potency in the 1b replicon assay.
Table 5: Compounds 11a-11s.
Such inhibition properties could be improved by the introduction of a 3-thiophene methylene R2 group (12y). Collectively, the above results demonstrate that numerous aliphatic moieties tethered to the pyridazinone N-2 atom by a methylene group could serve as effective R2 substituents for this series of NS5B inhibitors. In general, the pyridazinone-containing NS5B inhibitors described in this work displayed somewhat weaker inhibition activities when tested against genotype 1a HCV polymerase than that against genotype 1b enzyme. The difference in these inhibition levels was within sixfold for most compounds, although larger and smaller variations were noted. All of the smaller deviations were detected during the exploration of the pyridazinone R1 moieties (11i, 11m, 11p, and 11s) while the larger differences were mostly observed during variation of the R2 substituents (12g, 12l, 12n–o, 12r–s, and 12u–x). The described pyridazinone-containing molecules have good potential to inhibit HCV polymerases derived from both 1a and 1b genotypes [18].
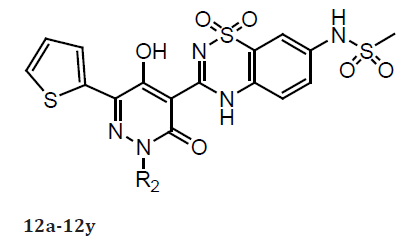
(Table 6) However, the data also caution that the selection of truly optimal inhibitor substituents, particularly at the pyridazinone R2 position, should be made using inhibition values obtained from both genotype 1a and 1b enzymes. In addition, the antiviral potencies (EC50) of the inhibitors under study measured against genotype 1b HCV replicon were usually weaker (2- to 20-fold) than the corresponding genotype 1b NS5B IC50 values. However, several exceptions to this general trend were noted (compounds 10m, 11l-11m, 11p, 12b-12d, 12g-12h, and 12p). The differences in the EC50/IC50 ratios are likely caused by variations in compound cell permeability and/or protein binding properties. The stability of many pyridazinone-containing inhibitors toward human liver microsomes was also assessed, and most compounds tested exhibited moderate to long half-lives (t1/2>30 min). Notable exceptions included molecules in which the R3 sulfonamide NH was alkylated or removed (10i-10q) and the inhibitors bearing alkyl or alkenyl R1 moieties (11g-11s).
Table 6: Compounds 12a -12y.
Importantly, many examples of pyridazinone-containing compounds were identified that combined potent NS5B inhibition properties with good stability toward human liver microsomes. Encouraged by the above results, we examined the pharmacokinetic properties of two very potent and metabolically stable NS5B inhibitors (10a and 12a) in monkeys after both oral (po) and intravenous administration of a 5 mg/kg single dose. As expected based on its good in vitro stability toward monkey liver microsomes (MLM), compound 10a exhibited a low in vivo clearance value after iv administration. Somewhat surprisingly, 12a displayed similar in vivo clearance properties in spite of its reduced MLM stability relative to 10a. These results suggested that the in vivo clearance of one or both compounds may not be primarily affected by CYPmediated biotransformation. Both inhibitors also exhibited low distribution volumes in the i.v experiments. Unfortunately, the bio availabilities of both compounds (10a and 12a) after oral administration to cynomolgus monkeys were very low. Since the molecules displayed low in vivo clearance values and reasonable solubilities in H2O at pH 7.4, we believe that their undesirable oral PK properties primarily result from poor gut permeability. Accordingly, both inhibitors displayed very low Papp values in Caco-2 permeability assessments.
These poor permeability properties are consistent with the highly polar nature of both 10a and 12a (polar surface area (PSA) = 203 A˚ 2 for both compounds) which places them considerably outside the polarity range typically associated with gut-permeable molecules (PSA<140 A˚2). In addition, Papp BA/AB ratios for both compounds 10a and 12a were very high suggesting that the low oral bio availabilities may be partially due to their ability to serve as efficient efflux substrates. In spite of the low oral bio availabilities exhibited by 10a and 12a, we found that the plasma concentrations of both compounds 12 h after oral administration to monkeys were close to their corresponding antiviral EC50 values [18]. As the liver is a major site of HCV replication, study revealed that the concentration of 3a in rat liver after oral administration was considerably higher than that in plasma for all time points examined. The oral liver/plasma (L/P) ratios declined over time, but consistent with the liver being the organ initially exposed to orally dosed agents, remained higher than the corresponding L/P ratios observed 12 h after iv administration of 10a. Prominently, the concentration of 10a in rat liver 12 h after po administration substantially exceeded its antiviral EC50 value (C12h/EC50 = 24). The high L/P ratio (64:1 12 h post oral dose) implied the tendency of 10a to accumulate at a major site of HCV replication and this property may enhance its ability to function as an effective anti- HCV agent in vivo [18].
A number of highly potent inhibitors of genotype 1 HCV NS5B polymerase, with single digit nM antiviral potency, derived from pyridazinones with a methyl sulfonamide R3 substituent. SAR studies revealed that R1 and R3 substituents considerably affected the HLM stability of the resulting molecules. A PK study in monkeys with the highly potent inhibitor 12a (1b EC50= 6 nM) using a single 5mg/kg oral dose showed poor oral bioavailability, but C12h in plasma was close to its EC50 value. The ratio of liver to plasma concentrations in rat of another very potent inhibitor 10a (1b EC50=5 nM) 12 h after a single oral dose of 3 mg/kg was very high (64-fold), and this property may be beneficial since liver is a major site of HCV replication [18].
The discovery and structure guided optimization of a series of 5-hydroxy-3(2H)-pyridazinone-containing NS5B inhibitors that bind to the ‘palm’ site. As part of these efforts, compound 16a was identified as one of the most potent molecules in this series with low nanomolar inhibition activity in both enzymatic (genotypes 1a and 1b) and cell-based (1b replicon) assays. Interestingly, compound 16a accumulated to relatively high levels in rat liver following oral dosing. However, 16a also exhibited very low bioavailability and low plasma exposures when orally administered to both rats and cynomolgus monkeys. Although we believe that the accumulation of 16a at a major site of HCV replication could facilitate its use as an anti-HCV agent, we also wished to identify other compounds that afforded more robust plasma exposures following their oral administration. We therefore pursued the further optimization of this series of pyridazinone-containing inhibitors in an effort to improve oral bioavailability [19].
Table 7: Compounds 16- 16a.
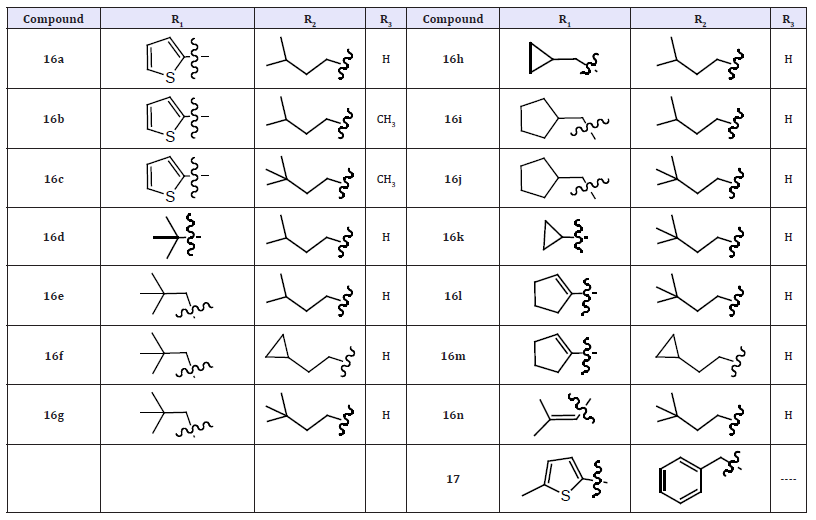
(Table 7)

(Table 8) The low oral bioavailability exhibited by 16a was likely due to poor intestinal absorption caused by its highly polar nature and/or its ability to serve as an efficient efflux substrate. A metabolically stable control compound (17) whose calculated polar surface area (PSA) was much closer to the range typically associated with the well-absorbed molecules (PSA<140 A˚2). This compound displayed significantly improved Caco-2 permeability relative to 16a, a reduced potential for efflux, and increased oral bioavailability in monkeys. The improvement in oral bioavailability has significant increase in the fraction of 17 that was compared with 16a (FApo). The pyridazinone optimization strategy on identifying potent NS5B inhibitors with reduced PSA values relative to that of compound 16a.
Table 8: Compounds 17a-17m.
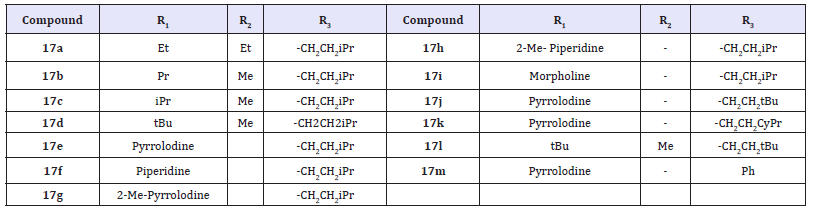
The molecules that incorporated R1, R2, and R3 fragments that were (16) less polar than those present in 16a and (17) known from our previous work to impart potent anti-NS5B properties against both genotypes 1a and 1b polymerases. To further differentiate designs with identical PSA values, we also calculated the corresponding fraction absorbed parameter (cFA) using a recently-developed computational algorithm that afforded good correlation with experimental fraction absorbed for a large number of existing drugs. The described strategy offered a good opportunity to more optimally balance the overall biological profiles of the inhibitors. N-methylation of the terminal sulfonamide present in 1a afforded improvements in Caco-2 permeability, efflux ratio, and in vivo fraction absorbed (FApo = 33%) (compare compound 16b with 16a).
These improvements were realized at the expense of metabolic stability (in addition to loss of antiviral potency), and this detriment likely attenuated the extent of increase in observed oral bioavailability as compared with 16a. Interestingly, replacing the isoamyl R2 moiety present in 16b with a tert-butyl ethyl group worsened the corresponding oral bioavailability of the resulting compound 16c. Consistent with this result, compound 16c exhibited significantly lower Papp and FApo values suggesting that poor absorption was responsible for the observed bioavailability differences. However, since the chemical structures and calculated PSA and cFA values of these two inhibitors are quite similar. Reduction in the PSA values of the compounds under study was achieved by replacing the R1 thiophene moiety present in 16a–16c with alkyl or alkenyl groups (16d–16n). Not unexpectedly, the increased lipophilicities of these molecules almost always afforded increased Papp values and reduced HLM/MLM stabilities relative to 16a. One exception was compound 1f that exhibited t1/2 > 60 min in the MLM assay (compare 16f with 16e) [19].
Unfortunately, the majority of these compounds also exhibited minimal improvements relative to 16a in both in vivo fraction absorbed (FApo) and bioavailability properties (Fpo) following oral administration to monkeys. Notable exceptions included inhibitors 16d, 16i, and 16j each of which displayed improved in vivo bioavailability and in vivo FApo values as compared with the lead molecule (16a). In addition to lower PSA properties, all these compounds had significantly higher cFA and Papp values as compared with 16a. While we believe that these differences likely contributed to their improved oral bio availabilities, related improvements in bioavailability were not observed for compounds 16k and 16n which also possessed similarly favorable PSA, cFA and even higher Papp values. All these compounds exhibited low to moderate clearance and low steady-state volumes of distribution (<1 L/kg) with moderate to good antiviral potency (16b EC50 = 110- 320 nM).
Although these molecules showed an improvement in both oral bioavailability and Cmax parameters as compared with 16a, they exhibited significantly lower C12h/EC50 ratios. In addition, MLM stability for these compounds did not correlate well with the corresponding clearance data suggesting that this process may not be primarily mediated via biotransformation. Collectively, these results illustrate the challenges we faced in optimizing the DMPK properties of the molecules in this series. In order to further improve the oral bioavailability and retain the antiviral potency of this series of pyridazinone compounds, we performed several analyses to help identify which calculated parameter, measured property, or combination thereof conferred the beneficial in vivo performance to the molecules. The molecules that possessed lower PSA values had a greater chance of exhibiting Caco-2 permeabilities in excess of the low control.
The more permeable compounds (Papp > low control), in turn, had an increased likelihood of displaying in vivo FApo values above 10%. Collectively, this analysis confirmed that reduction in PSA values could favorably impact the permeability properties of the pyridazinone-containing NS5B inhibitors described in this work. However, for compounds with identical PSA characteristics (e.g., 174 A˚2), the in vivo fraction absorbed (FApo) varied from 2% to 18%. In addition, there was little correlation noted between log D values and the in vivo fraction absorbed. Consistent with this observation, the calculated fraction absorbed (cFA) values (which were based on both calculated PSA and calculated log D) associated with the above compounds showed little to no correlation with the corresponding experimental fraction absorbed parameters. A number of potent inhibitors of genotype 1 HCV NS5B polymerase derived from pyridazinones were evaluated in both in vitro assays and in vivo animal PK studies. The described optimization focused on DMPK properties and led to potent inhibitors (16b EC50 = 110– 320 nM) with improved oral bio availabilities (10–15%) relative to the lead compound (1a, 2%). In addition, analysis of the correlation between calculated parameters, in vitro biological properties, and the measured fraction absorbed or oral bioavailability in animals defined relationships that could be applied in the further optimization of the DMPK properties of the molecules in this series [19].
We previously disclosed the identification of a novel series of non-nucleoside NS5B inhibitors containing a 1,1-dioxo- 1,4-dihydro-1k6-benzo [1,2,4] thiadiazine moiety linked to a substituted 5-hydroxy-3(2H)-pyridazinone. Many such compounds exhibit potent NS5B inhibition activities and often display submicromolar antiviral properties when tested against an HCV replicon in cell culture. Unfortunately, these molecules also frequently exhibit poor pharmacokinetic properties following oral administration to animals. In an effort to improve these undesirable pharmacokinetic characteristics, we sought to diversify the 5-hydroxy-3(2H)-pyridazinone pharmacophore by the inclusion of a 6-amino-substituent into the inhibitor design. The desired 6-aminopyridazinones (19) were prepared using two related synthetic methods. Each of these methods involved derivatization of an appropriately substituted 6-amino-5- hydroxy-3(2H)- pyridazinone intermediate bearing an ethyl ester moiety at the 4-position (20) [20].

Many of these molecules incorporate an isoamyl (CH2CH2iPr) substituent at the pyridazinone 2-position (R3 in structure 19) that was known from our earlier studies to impart potent NS5B inhibition properties to pyridazinone-containing compounds. For example, a molecule that contained such an R3 fragment and a diethylamino moiety at the 6-position on the pyridazinone ring displayed submicromolar levels of both NS5B inhibition activity and antiviral potency (17a). Several related compounds bearing N-methyl-Nalkyl- aminopyridazinone 6-substituents were also examined, and the corresponding NS5B inhibition properties were observed to improve somewhat for those containing bulkier alkyl groups (17b with 17c and 17d). An even more dramatic improvement in both anti-NS5B and antiviral activity was realized by replacing the diethylamino moiety present in the lead molecule with a pyrrolidine ring (17e with 17a). However, incorporating a piperidine moiety at this location did not afford similar enhancements (17f with 17a). These results parallel the SAR trends previously observed during our variation of alkyl and aryl pyridazinone 6-substituents in which compounds bearing 5-membered cyclic structures were more potent NS5B inhibitors than analogs containing 6-membered moieties. Interestingly, addition of a methyl group adjacent to the ring nitrogen in compounds 2e and 2f had opposite effects on inhibitor potency.
Derivatization of the former compound in this manner resulted in a worsening of anti-NS5B and antiviral properties (17e with 17g), while similar modification of the latter molecule slightly improved NS5B inhibition activity. Incorporation of a morpholine moiety in lieu of the piperidine ring in the inhibitor design provided a considerable improvement in anti-NS5B properties (17i with 17f). However, this alteration was detrimental to antiviral activity in cell culture, as 17i was considerably weaker in the replicon assay than several other compounds with equivalent NS5B inhibition properties (17i with 17e, 17j, and 17k). The large EC50/IC50 ratio exhibited by 17i is likely due to the compound’s poor cell membrane permeability properties. Consistent with this hypothesis, 17i exhibited a Caco-2 Papp value that was significantly lower (less permeable) than all of the other 6-aminopyridazinones examined in this work. Several additional NS5B inhibitors were prepared which combined optimal 6-amino substituents with non-isoamyl R3 moieties that were also known from our previous SAR studies to impart potent NS5B inhibition properties to the compounds that contained them. For example, a molecule incorporating an R3 tertbutyl ethyl (CH2CH2tBu) fragment and a pyrrolidine substituent at the 2-position of the pyridazinone ring displayed excellent NS5B inhibition activity and potent antiviral properties in cell culture (17j). Similarly, potent anti-NS5B characteristics were noted for a compound which contained an R3 cyclopropyl ethyl moiety (CH2CH2CyPr) in lieu of the tert-butyl ethyl fragment (17k).
However, the antiviral activity exhibited by the latter molecule was significantly worse than that displayed by the former inhibitor. This difference may be due to differing cell membrane permeability properties of the compounds in question, although the corresponding Caco-2 Papp values were similar for both molecules. Somewhat surprisingly, combination of the tert-butyl ethyl R3 fragment with a N-methyl-N-tert-butyl-aminopyridazinone 6-substituent resulted in somewhat poorer NS5B inhibition activity relative to the corresponding R3 isoamyl analog (17l with 17d). However, the two compounds displayed similar antiviral properties in the cell-based assay. While the precise reasons for these activity trends are not currently understood, these results (along with those observed above for compounds 17g and 17h) suggest that NS5B recognition by this class of inhibitors is highly sensitive to subtle structural variations within both the R3 moiety and the 6-aminopyridazinone substituent. Accordingly, a molecule incorporating a phenyl R3 fragment (17m) displayed reduced NS5B inhibition activity and drastically attenuated antiviral properties relative to the most potent compounds described in this work [20].
The stability of the aminopyridazinone-containing compounds toward human liver microsomes (HLM) was also assessed to better predict their potential for metabolic transformation in vivo. These results suggest that incorporation of 6-amino-substituents into the pyridazinone-containing NS5B inhibitor series introduces at least one metabolically labile site into the inhibitor design. One exception to this trend was compound 17i which displayed a long half-life in the HLM assay. As described above, however, this molecule also exhibited reduced Caco-2 permeability and antiviral properties relative to the other aminopyridazinone-containing inhibitors. The other exception (compound 17m) displayed even poorer activity in the replicon cell-based assay. These observations highlight the difficulty of improving the HLM stability of this inhibitor series while simultaneously retaining good potency, permeability, and/or absorption characteristics.
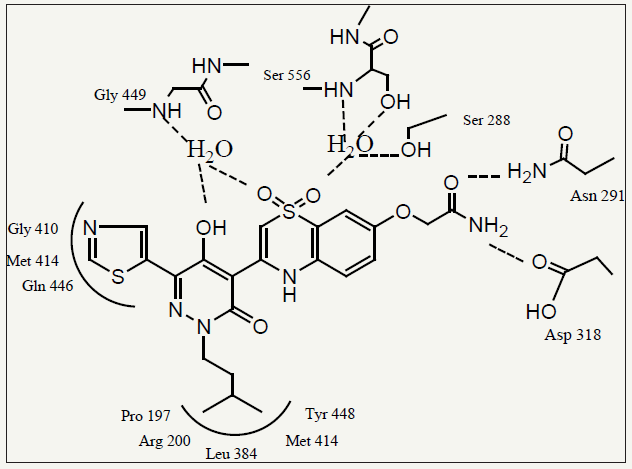
The X-ray crystal structure of compound 17e bound to the NS5B protein. A schematic diagram depicting enzyme residues near the inhibitor as well as protein–ligand hydrogen bonding interactions is also provided (Figure 3). As was observed in previous pyridazinone- NS5B crystal structures, the R3 isoamyl fragment of 17e bound in a deep hydrophobic pocket comprised of NS5B residues Pro- 197, Arg-200, Leu-384, Cys-366, Met-414, Tyr-415, and Tyr-448. In contrast, the pyrrolidine ring present in 17e bound to a much more shallow cleft near the surface of the enzyme that was made up of residues Gly-410, Met-414, and Gln-446. The 5-hydroxy group on the pyridazinone ring formed hydrogen bonds with the backbone amide NH of Tyr-448 and a structural water molecule that interacted with the backbone NH of Gly-449. In addition, one of the oxygen atoms present in the benzothiadiazine ring system of 17e and one of the methylsulfonamide oxygen atoms each formed a hydrogen bond with a second structural water molecule (itself hydrogen-bonded to the side chain of Ser-288) [20].
figure 3: Compound 17e bound in the NS5B palm site. Hydrogen bonds are represented as dashed lines, and the residues which make up the enzymebinding subsites are depicted.
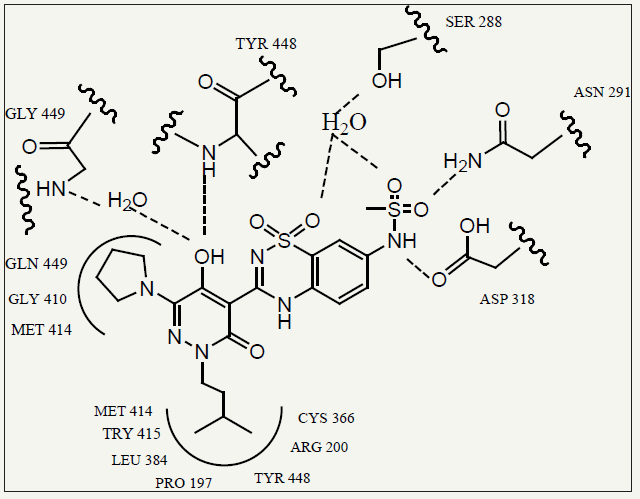
Hydrogen bonds were also noted between the other methylsulfonamide oxygen atom and the side chain of Asn-291 as well as between the methylsulfonamide NH moiety and the side chain of Asp-318. Similar hydrogen bonding interactions were observed in co-crystal structures of other benzothiadiazine containing NS5B inhibitors with NS5B, and these likely contribute to the potent polymerase inhibition properties exhibited by many of the compounds. The 4-(10,10-dioxo-10,40-dihydro-10k6- benzo[10,20,40]thiadiazin-30-yl)-5-hydroxy-2H-pyridazin-3- ones bearing 6-amino substituents as potent inhibitors of the HCV RNA-dependent RNA polymerase (NS5B). Many of these agents also display antiviral activity in cell culture experiments. However, in vitro DMPK data suggest that it may be difficult to combine potent antiviral activity, good metabolic stability, and favorable permeability/absorption characteristics in this class of NS5B inhibitors [20]. The 4-(1’,1’-dioxo-1’,4’-dihydro-1’lambda(6)- benzo[1’,2’,4’]thiadiazin-3’-yl)-5-hydroxy-2H-pyridazin-3-ones bearing 6-amino substituents as potent inhibitors of the HCV RdRp NS5B. Several of these agents also displayed potent antiviral activity. In vitro DMPK data (microsome t1/2, Caco-2 P(app)) for many of the compounds were disclosed, and a crystal structure of a representative inhibitor complexed with the NS5B protein was discussed [20].
Hepatitis C virus (HCV) is a leading cause of chronic liver disease. Current therapies for genotype 1 HCV are associated with sub-optimal response rates and debilitating side effects. The efforts to discover non-nucleoside small molecule inhibitors of genotype 1 HCV polymerase, we investigated a series of 5-hydroxy- 3(2H)-pyridazinones using a structure-based design approach. We systematically explored variation of the substituents located at the 2-, 4- and 6-positions on the pyridazinone ring. A number of the analogs we prepared were found to inhibit the NS5B enzyme
with low nanomolar potencies. The SAR observed by varying the 4-substituent present in our 5-hydroxy-3(2H)-pyridazinone NS5B inhibitors. Variation of this substituent has a dramatic impact on NS5B binding affinity. We identified the methylsulfonylaminosubstituted benzothiadiazine as among the best 4-substituents and its inclusion into our pyridazinones afforded NS5B inhibitors with low nanomolar potencies against both the HCV polymerase and replicon. Optimization of the 4-substituent present in a series of 5-hydroxy-3(2H)-pyridazinones provided NS5B inhibitors with low nanomolar potencies in both biochemical and HCV replicon assays. These inhibitors generally display reasonable solubility properties. Some analogs exhibit very high liver to plasma ratios after oral administration to rats [21]. Hepatitis C virus (HCV) is a leading cause of chronic liver disease. Current therapies for genotype 1 HCV are associated with sub-optimal response rates and debilitating side effects. The non-nucleoside small molecule inhibitors of genotype 1 HCV polymerase, a series of 5-hydroxy-3(2H)-pyridazinones (22) using a structure-based design approach. We systematically explored variation of the substituents located at the 2-, 4- and 6-positions on the pyridazinone ring. A number of the analogs were found to inhibit the NS5B enzyme with low nanomolar potencies [22].
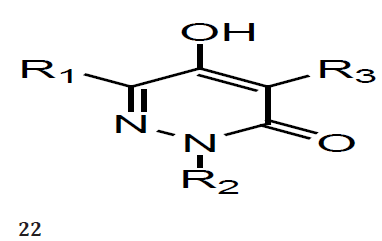
Described here are the SAR observed by varying the 2- and 6-substituents of our 5-hydroxy-3(2H)-pyridazinone NS5B inhibitors. We observed that small heteroaromatic rings and alkyl groups were optimal 6- substituents. Certain 2-substituents improved enzyme inhibitory potency against genotype 1a NS5B. The combination of optimal substituents at positions 2 and 6 resulted in inhibitors with low nanomolar potencies against genotype 1a/1b NS5B enzymes and the genotype 1b HCV replicon. Optimization of the 2- and 6-substituents in a series of 5-hydroxy- 3(2H)-pyridazinones provided NS5B inhibitors with low nanomolar potencies in both biochemical and replicon assays. These inhibitors generally display reasonable solubility properties. Some analogs exhibit very high liver to plasma ratios after oral administration to rats [22].
The N-2 position of pyridazinone 23, a potent HIV-1 NNRTI that has limited aqueous solubility, was derivatized into a series of hydroxymethyl esters and carbonates as well as one phosphate and served as prodrugs to effectively deliver 1 to rat plasma upon oral treatment at 50 mpk. Increases of 4.3- to 8.6-fold in 24-hour exposure of 23 (over that of parent) were observed while the prodrugs and the hydroxymethyl adduct 24 were undetectable. The SAR of HIV-1 reverse transcriptase inhibitors compound 1 is a representative. These pyridazinones display high potency and bind at the allosteric site common to most non-nucleoside inhibitors (NNRTI). Compound 1 has very good Caco2 permeability and stability in human liver microsomes as well as low iv clearance rates in rat and dog, 16% and 14% of hepatic blood flow, respectively. However, 1 displays poor aqueous solubility and a high melting point, both of which are typical characteristics for the chemotype. Plasma exposure was less than dose-proportional after oral administration with only a 3-fold increase in 24-hour AUC for the 10-fold increase in dose. These data for 5 and 50 mpk suggest that poor solubility and high crystallinity are playing significant roles to limit oral exposure. Furthermore, levels of 1 at the 50 mpk dose did not reach target plasma levels desired to provide a high antiviral inhibitory quotient of 1 that warranted its further development as an antiviral candidate. Prod rugs have been utilized to overcome delivery problems of drugs with limited solubility, for instance phenytoin. The prod rug fosphenytoin bears a solubilizing phosphate feature. The approach to generate a stable prod rug was achieved by linking that feature via a hydroxymethyl derivative. Phenytoin is a substituted hydantoin that is more acidic (calcd pKa 8) than an amide and this property enables a rapid release of the parent upon cleavage of the phosphoryl bond of fosphenytoin [23].
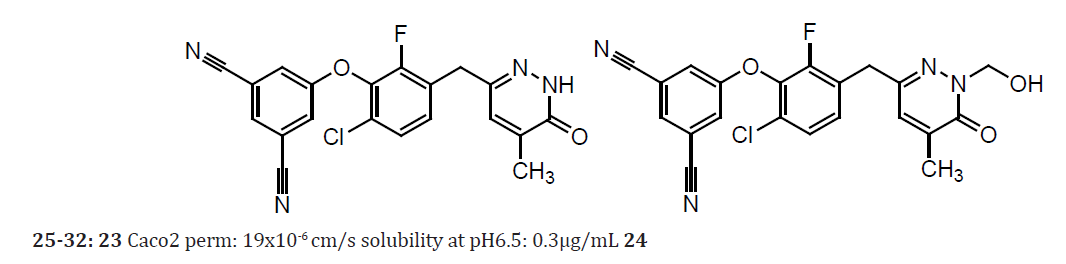
Table 9: Compound 25-32.
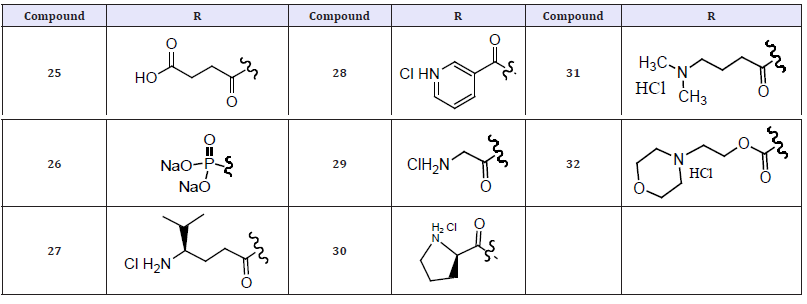
(Table 9) Inhibitor 23 displays acidity (pKa 10.5) similar to that of phenytoin thus a rapid in vivo release of parent was anticipated. This describes a successful approach to improve the blood plasma levels of 1. These prodrugs were anionic or cationic species so as to promote dissolution. Isopropenyl chloroformate served as an excellent activating agent (for acid substrates). The succinate half ester was generated as the free acid, while the nicotinate was evaluated as the hydrochloride salt. The requisite acid substrates for esters were the N-Boc amino acids of Gly and Pro, respectively. The esters were obtained as their HCl salts following the deprotection under acidic conditions. The phosphate ester was generated as its di-sodium salt. The S-Val derived N-carboxyanhydride readily consumed 24 at ambient temperature and ester was obtained upon acid treatment. The generation of carbonates by initial acylation of 2 with 4-nitrophenoxy chloroformate, followed by treatment with 2-(N,Ndimethylamino) ethanol or 4-(2-hydroxyethyl) morpholine to give 9 or 10, respectively [23].
Amino acid derivatives have reported to increase absorption by targeting oligopeptide transporters. Prodrugs were generated in interest of their salts to enhance dissolution. The prodrugs and adduct 2 were not detected in rat plasma during the 24-hour course of the pharmacokinetic evaluations. Pyridazinone 1 was found to be an excellent substrate to derivatize as hydroxymethyl linked prodrugs that in turn gave rapid in vivo liberation of parent. Substantially improved plasma levels of 1 were observed following the oral administration of prod rugs to rat. Phosphate prod rug presented superior aqueous stability and the most efficient delivery of 25. An 8.6-fold enhancement of the circulating NNRTI 25 was observed during the 24-hour exposure. In contrast to previous experience with nucleoside-amino ester prodrugs, prodrugs of 1 bearing a lipophilic amino ester do not appear to be substrates for active transport, they did not enhance bioavailability relative to the other prodrugs. Despite differences in aqueous stability, consistent and robust increases in 24-hour exposure, from 4.3- to 6.9-fold were obtained across the series of acyl prodrugs examined. Plasma concentrations of prodrugs were undetectable thus apparently consistent with prodrug conversion by ubiquitous esterase activity. Therefore, the proposal we forward is that the acyl cleavage of the prodrugs is initially subject to enzymatic action during absorption that likely was not saturated under the study conditions [23].
The identification of compound 25 from a high throughput screen directed against the NS5B polymerase. In a secondary replicon assay4this compound displayed moderate activity against HCV genotype 1b (EC50=2lM) but was 10-fold less active (EC=25l M) in a related HCV genotype 1a assay. Exploratory lead optimization led to the identification of significantly more potent analogs such as compound 26. This molecule displayed EC values in the replicon 1b and 1a assays of 7 nM and 87 nM, respectively. A notable feature of the SARs observed during the early development of this series was the consistently enhanced activity against genotype 1b, despite the introduction of significant structural modification [24, 25].
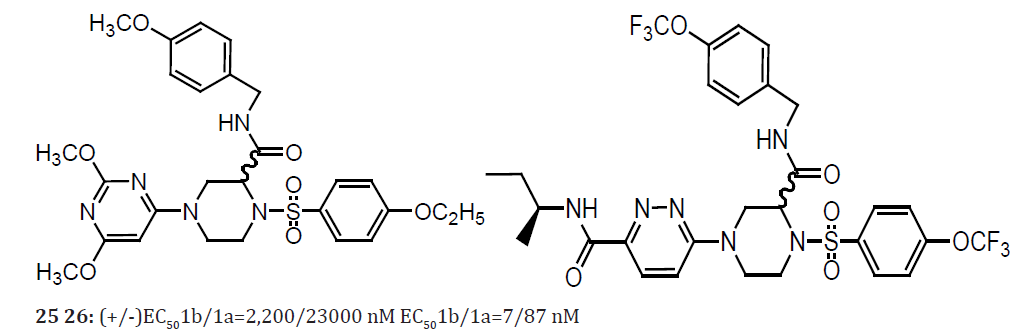
Piperazine based HCV NS5B inhibitors displaying significant selectivity for genotype 1b over genotype 1a. Compound 25 is racemic, and compound 26 has the absolute stereochemistry shown. The select co-crystal structures were obtained with analogs from this series complexed with the NS5B protein. The structural data proved to be particularly helpful in explaining many aspects of the observed SAR. However, these data were less helpful in identifying specific interactions between the ligands and the NS5B 1b and 1a proteins that could explain the observed genotype selectivity, as discussed below. In addition to compounds 25 and 26, co-crystal structures with NS5B were obtained with the analogs 3, 4 and 5 depicted. The syntheses of all of these compounds have been previously reported, and all are accessible by relatively concise synthetic routes. This pocket is exposed by a movement of the C-terminus that allows entry to a cavity that is otherwise inaccessible in the apo structure. Interestingly, the 4-ethylbenzenesulfonamide moiety binds in a hydrophobic pocket which is occupied by residues W550 and F551 in the NS5B apo structure (1C2P89 in the Protein Data Bank).
These two residues are part of a C-terminal regulatory motif which modulates the activity of NS5B. With respect to specific ligand–protein interactions, the pyrimidine ring of 25 participates in a p -stacking interaction with F193, and the methoxyphenyl ring p-stacks with Y448. The carboxamide moiety of the ligand donates an H-bond to the Y195 backbone carbonyl, and the guanidine moiety of R200 is proximal to the methoxyphenyl ring in the ligand with an NH directed towards the p -face of the ring suggesting a cation– p interaction. The structure suggests that the (R)-stereochemistry is required at the chiral center on the piperazine ring, which was confirmed by synthesis using specific piperazine enantiomers. The functionalities projecting from the carboxamide and sulfonamide vectors in 1 are already highly complementary to the cavities into which they project. Consistent with this observation was the finding that attempts to incorporate significant structural modification at either of these vectors resulted in a notable loss of activity.
Despite testing an extensive series of analogs, only modest gains in potency could be achieved, as observed with piperazine. This analog displayed EC values against genotypes 1b and 1a of 700 nM and 8100 nM, respectively. A co-crystal structure of compound 3 complexed with NS5B genotype 1b was obtained. Piperazine 3 was found to bind in a similar fashion to compound 1. Despite the lower resolution of this structure more residues of the NS5B C-terminus were resolved, and L547, W550, F551, Y555, and S556 are proximal to the bound ligand. A hydrogen bond between a methoxy group on the ligand pyrimidine ring and the backbone NH of G449 is also observed. However, as aromatic ether oxygens are poor hydrogen bond acceptors, this is likely to contribute little to the potency. Slightly more extensive vander Waals interactions between the trifluoromethoxy group and the hydrophobic pocket into which the sulfonamide moiety projects probably account for the modest potency improvement observed with this analog relative to 25.
A notable feature in both complexes is that a significant unoccupied volume exists around the pendant pyrimidine moiety, the SAR favors pyridazine heterocycles at this position. In order to understand this, a co-crystal structure was obtained with the pyridazine ligand 4 complexed to NS5B genotype 1b. Several interesting features in this structure were observed. First, the principal hydrophobic interactions exhibited by the pendant sulfonamide and carboxamide moieties are retained, and the latter maintains a hydrogen bond with the Y195 backbone carbonyl. Second, the pyridazine ring still appears to participate in a p-stacking interaction with F193.The observed EC50 values for 4 of 700 nM and 2200 nM against genotypes 1b and 1a, respectively. It is apparent from the structure depicted that 3,6-disubstituted pyridazines with relatively small substituents could be accommodated and may be associated with enhanced activity. A SAR study was conducted to determine the optimal moiety at the 6-position of the heterocycle and involved the preparation of a focused array of N-alkyl substituted carboxamides.
This substitution was associated with a dramatic enhancement of potency, as observed with compound 26 that displayed EC values against genotypes 1b and 1a of 7 n M and 87 nM, respectively. A co-crystal structure of 25 complexed with NS5B genotype 1b was obtained. In addition to the previously mentioned interactions, a number of other binding contacts were observed, the most significant being H-bonding between Y555 and S288 and the 6-carboxamide moiety, and additional hydrophobic contacts with the carboxamide N-alkyl substituent. One of the pyridazine ring nitro- gen atoms also forms water-bridged hydrogen bonds with the protein. Collectively, these interactions could account for the enhanced potency observed for compound 26. With potent analogs identified, the origin of the consistent 1b genotype selectivity exhibited by this compound class was explored. The most significant differences between the structures occur in the vicinity of the carboxamide moiety.
However, careful analysis of the geometrically non-optimal H-bonding arrays in this area suggests these differences are unlikely to account for the genotype selectivity exhibited. Only a single amino acid difference exists between genotypes 1b and 1a in the region proximal to that occupied by the ligand; a tyrosine is present at position 415 in NS5B 1b, and phenylalanine occupies this position in genotype 1a. It is difficult to rationalize how this sequence change can account for the potency differences observed. Given that this binding site is accessible only on displacement of the C-terminus, it is possible that the dynamics of the NS5B 1b and 1a proteins are different and could account for the observed potency differences. Support for this possibility is provided by the fact that part of the binding site for the ligands is occupied by W550 and F551 of the C-terminal regulatory motif in the Apo structure of NS5B 1b.
Along with L447, these residues serve to anchor the C-terminus to the surface of the protein. Interestingly, the two residues flanking W550 and F551 differ in genotypes 1a and 1b (1a: G549, T552; 1b: S549, V552). It may be that these sequence differences modulate the propensity of the C-terminus for displacement or alter the conformational preferences of the C-terminus once it dissociates from the binding site. Alternatively, crystal packing in the two different crystal forms may account for the observed positions of the mobile C-terminal region which may not represent solution conformations. Another possibility is that some interaction(s), as yet unspecified, remote from the binding site may be responsible for the diminished activity observed with the 1a genotype. The structures described rationalize much of the SAR observed with the piperazine class of NS5B inhibitors, although no clear explanation for the observed genotype selectivity has yet emerged [24,25].
Discussion
The Current therapies for HCV are based on combinations of pegylated interferon (IFN-a) and ribavarin, a broad spectrum antiviral agent. In addition, it is apparent that further optimization of the pendant heteroaryl-carboxamide moiety may be merited, and it would seem reasonable to anticipate improvements in potency that may be sufficient to accommodate the reduced level of activity consistently observed with NS5B 1a genotype. The optimization of (+/-) N-benzyl-4-heteroaryl-1-(phenylsulfonyl) piperazine-2-carboxamide against the NS5B polymerase enzyme of the hepatitis C virus. The identification of (S)-N-sec-butyl-6-((R)-3- (4-(trifluoromethoxy) benzylcarbamoyl)-4-(4-(trifluoromethoxy) phenylsulfonyl) piperazin-1-yl) pyridazine-3-carboxamide as a potent replicon activities against HCV genotypes 1b and 1a [24,25].
The NS5B is an RNA-dependent RNA polymerase that plays a key role in the infection cycle of hepatitis C. The HCV is a RNA virus of the Flaviviridae family. Chronic hepatitis C can lead to cirrhosis and end-stage liver disease in 20-30% of the patients, among these 1-4 % may develop hepatocellular carcinoma. The currently therapeutic protocol includes a combination of pegylated interferon and ribavirin [26-30]. However, these regimens have limited efficacy (10-40% of the patients) and significant side effects, causing up to 20% of the patients to discontinue the therapy. There is an urgent need for developing safe and effective antiviral agents. In the last decade the development of inhibitors targeting the HCV NS5B polymerase (RdRp) has attracted the attention of investigators worldwide. Specific inhibitors for HCV RdRp were however not identified until recently. These include nucleoside analogues and various non-nucleosides from different chemical classes.
The HCV infection is a blood-born disease that targets the liver and progresses to organ cirrhosis and primary cancer in a significant proportion of patients. The currently available treatment has limited efficacy and suffers from restricting side effects. Virus-specific, more efficacious, and better-tolerated anti- HCV therapies are thus required to address the unmet medical need. The development of HCV NS5B polymerase inhibitors are as potential therapy for the treatment of HCV infection. Several molecule inhibitors of HCV NS5B have progressed into clinical development and demonstrated efficacy in reducing viral load in infected patients. The Hepatitis C virus (HCV) is the main agent responsible for contagious hepatitis not due to the “classical” hepatitis viruses A and B. There is presently no vaccine or widely effective treatment for the resulting illness, which was previously called non-A-non-B hepatitis. HCV infection has been recognized as an urgent health crisis primarily. Because the virus causes few acute symptoms, most HCV patients are unaware of their infection.
However, if left untreated, the majority of all HCV infections lead to chronic hepatitis, which can develop into cirrhosis and liver cancer. Because current HCV therapies are costly and produced ebilitating side effects, new antiviral therapies are clearly needed. HCV antiviral drug development has been difficult because the virus cannot be conventionally cultivated in tissue culture or a convenient animal model. Consequently, the viral replicase has been intensely targeted for rational drug design. Like the majority of viruses, HCV does not use DNA to store its genetic material. Instead, the RNA genome of HCV is translated by host ribosomes directly into viral structural components and the machinery required for viral replication. This “replicase” is composed of several proteins that work together to produce new viral particles from the RNA genome. The two components that are best characterized are the NS5B RNA-dependent RNA polymerase and the NS3 protease/helicase. This review discusses progress in understanding the mechanism of action of these proteins. Consequently, the compounds interfere with the life cycle of the hepatitis C virus and are also useful as antiviral agents. The present invention relates to a subject suffering from HCV infection. The invention relates to treating an HCV infection [30-33].
Conclusion
Current therapies for HCV are based on combinations of pegylated interferon (IFN-a) and ribavarin, a broad spectrum antiviral agent. Therefore, the discovery of specific antiviral agents would be of great benefit to HCV patients. So, the need is to an effort to discover novel anti-HCV agents, which are targeted on two essential virally-encoded enzymes, NS3 serine protease and NS5B polymerase in this drug discovery and development effort.
References
- Assis DN, Lim JK (2012) New pharmacotherapy for hepatitis C. Clin Pharmacol Ther 92(3): 294-305.
- Barreca ML, Iraci N, Manfroni G, Cecchetti V (2011) Allosteric inhibition of the hepatitis C virus NS5B polymerase: in silico strategies for drug discovery and development. Future Med Chem 3(8): 1027-1055.
- Baugh J, Gallay P (2012) Cyclophilin involvement in the replication of hepatitis C virus and other viruses. Biol Chem 393(7): 579-587.
- Belda O, Targett-Adams P (2012) Small molecule inhibitors of the hepatitis C virus-encoded NS5A protein. Virus Res 170(1-2): 1-14.
- Davis BC, Thorpe IF (2013) Thumb inhibitor binding eliminates functionally important dynamics in the hepatitis C virus RNA polymerase. Proteins 81(1): 40-52.
- Eberle J, Gürtler L (2012) The evolution of drug resistance interpretation algorithms: ANRS, REGA and extension of resistance analysis to HIV-1 group O and HIV-2. Intervirology 55(2): 128-133.
- Gane EJ, Stedman CA, Hyland RH, Ding X, Svarovskaia E, et al. (2013) Nucleotide polymerase inhibitor sofosbuvir plus ribavirin for hepatitis C. N Engl J Med 368(1): 34-44.
- Gerber L, Welzel TM, Zeuzem S (2013) New therapeutic strategies in HCV: polymerase inhibitors. Liver Int 33(Suppl1): 85-92.
- Hunt D, Pockros P (2013) What are the promising new therapies in the field of chronic hepatitis C after the first-generation direct-acting antivirals? Curr Gastroenterol Rep 15(1): 303.
- Ismail MA, Abou El Ella DA, Abouzid KA, Mahmoud AH (2012) Integrated structure-based activity prediction model of benzothiadiazines on various genotypes of HCV NS5b polymerase (1a, 1b and 4) and its application in the discovery of new derivatives. Bioorg Med Chem 20(7): 2455-2478.
- Kiser JJ, Flexner C (2013) Direct-acting antiviral agents for hepatitis C virus infection. Annu Rev Pharmacol Toxicol 53: 427-449.
- Mahmoud AH, Abou El Ella DA, Ismail MA, Abouzid KA (2012) A highly selective structure-based virtual screening model of Palm I allosteric inhibitors of HCV Ns5b polymerase enzyme and its application in the discovery and optimization of new analogues. Eur J Med Chem 57: 468- 482.
- Pawłowska M (2011) Attempts to treat chronic hepatitis C with HCV protease inhibitor. Przegl Epidemiol 65(1): 35-38.
- Rong L, Ribeiro RM, Perelson AS (2012) Modeling quasispecies and drug resistance in hepatitis C patients treated with a protease inhibitor. Bull Math Bio 74(8): 1789-1817.
- Saeed M, Scheel TK, Gottwein JM, Marukian S, Dustin LB, et al. (2012) Efficient replication of genotype 3a and 4a hepatitis C virus replicons in human hepatoma cells. Antimicrob Agents Chemother 56(10): 5365- 5373.
- Zhou Y, Li LS, Dragovich PS, Murphy DE, Tran CV, et al. (2008) Novel HCV NS5B polymerase inhibitors derived from4-(1’,1’-dioxo-1’,4’-dihydro- 1’λ6-benzo[1’,2’,4’]thiadiazin-3’-yl)-5-hydroxy-2H-pyridazin-3-ones: Part 2: Variation of the 2- and 6-pyridazinone substituents. Bioorg & Med Chem Lett 18(4): 1419-1424.
- Zhou Y, Webber SE, Murphy DE, Li LS, Dragovich PS, et al. (2008) Novel HCV NS5B polymerase inhibitors derived from 4-(1’,1’-dioxo-1’,4’- dihydro-1’k6-benzo[1’,2’,4’]thiadiazin-3’-yl)-5-hydroxy-2H-pyridazin-3- ones. Part 1: Exploration of 7’-substitution of benzothiadiazine. Bioorg & Med Chem Lett 18: 1413-1418.
- Li LS, Zhou Y, Murphy DE, Stankovic N, Zhao J, et al. (2008) Novel HCV NS5B polymerase inhibitors derived from 4-(1’,1’-dioxo-1’,’dihydro- 1’k6-benzo[1’,2’,4’] thiadiazin-3’-yl)-5-hydroxy-2H-pyridazin-3-ones: Part 3: Further optimization of the 2-, 6-, and 7’-substituents and initial pharmacokinetic assessments. Bioorg & Med Chem Lett 18(11): 3446- 3455.
- Sergeeva MV, Zhou Y, Bartkowski DM, Nolan TG, Norris DA, et al. (2008) Novel HCV NS5B polymerase inhibitors derived from 4-(1’,1’-dioxo-1’,4’- dihydro-1’k6-benzo[1’,2’,4’] thiadiazin-3’-yl)-5-hydroxy-2H-pyridazin- 3-ones: Part 4: Optimization of DMPK properties. Bioorg & Med Chem Lett 18(11): 3421-3426.
- Dragovich PS, Blazel JK, Ellis DA, Han Q, Kamran R, et al. (2008) Novel HCV NS5B polymerase inhibitors derived from 4-(1’,1’-dioxo-1’,4’- dihydro-1’lambda(6)-benzo[1’,2’,4’]thiadiazin-3’-yl)-5-hydroxy-2Hpyridazin- 3-ones: Part 5: Exploration of pyridazinones containing 6-amino substituents. Bioorg Med Chem Lett 18(20): 5635-5639.
- Zhou Y, Li LS, Webber S, Dragovich P, Murphy D, et al. (2007) Potent HCV NS5B Polymerase Inhibitors Derived From 5-Hydroxy-3(2H)- Pyridazinones: Part 1 Exploration of Pyridazinone 4-Substituent Variation. Program and Abstracts/Antiviral Research, Anadys Pharmaceuticals, Inc., San Diego, CA, USA, p. 97.
- Zhou Y, Li LS, Webber S, Ayida B, Bertolini T, et al. (2007) Potent HCV NS5B Polymerase Inhibitors Derived From 5-Hydroxy-3(2H)-Pyridazinones: Part 2: Variation of the 2- and 6-Pyridazinone Substituents. Program and Abstracts/Antiviral Research 74(3): A51-A52.
- Elworthy TR, Dunn JP, Hogg JH, Lam G, Saito YD (2008) Orally bioavailable prodrugs of a BCS class 2 molecule, an inhibitor of HIV-1 reverse transcriptase. Bioorg Med Chem Lett 18(24): 6344-6347.
- Gentles RG, Ding M, Zheng X, Chupak L, Poss MA (2011) SAR studies on a series of N-benzyl-4-heteroaryl-1-(phenylsulfonyl)piperazine-2- carboxamides: potent inhibitors of the polymerase enzyme (NS5B) of the hepatitis C virus. Bioorg Med Chem Lett 21(10): 3142-3147.
- Gentles RG, Sheriff S, Beno BR, Wan C, Kish K, Et al. (2011) Investigation of the mode of binding of a novel series of N -benzyl-4-heteroaryl- 1-(phenylsulfonyl)piperazine-2-carboxamides to the hepatitis C virus polymerase. Bioorg & Med Chem Lett 21(8): 2212-2215.
- Frick DN (2004) The Hepatitis C Virus Replicase: Insights into RNAdependent RNA Replication and Prospects for Rational Drug Design. Curr Org Chem 8(3): 223-241.
- Kronenberger B, Zeuzem S (2012) New developments in HCV therapy. J Viral Hepat (Suppl 1): 48-51.
- Lee LY, Tong CY, Wong T, Wilkinson M (2012) New therapies for chronic hepatitis C infection: a systematic review of evidence from clinical trials. Int J Clin Pract 66(4): 342-355.
- Neumann Haefelin C, Blum HE, Thimme R (2012) Direct antiviral treatment strategies in chronic hepatitis C. Dtsch Med Wochenschr 137(25-26): 1360-1365.
- Treviño A, De Mendoza C, Parra P, Rodríguez C, Madejón A (2011) Natural polymorphisms associated with resistance to new antivirals against HCV in newly diagnosed HIV-HCV-coinfected patients. Antivir Ther 16(3): 413-416.
- Waheed Y, Bhatti A, Ashraf M (2013) RNA dependent RNA polymerase of HCV: A potential target for the development of antiviral drugs. Infect Genet Evol 14: 247-257.
- Yang PL, Gao M, Lin K, Liu Q, Villareal VA (2011) Anti-HCV drugs in the pipeline. Curr Opin Virol 1(6): 607-616.
- Zhang HW, Zhou L, Coats SJ, McBrayer TR, Tharnish PM, et al. (2011) Synthesis of Purine Modified 2’-C -Methyl Nucleosides as Potential Anti- HCV Agents. Bioorg Med Chem Lett 21(22): 6788-6792.
© 2018 Mohammad Asif. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






