- Submissions

Full Text
Modern Approaches in Drug Designing
PARP Inhibitors: A Breakthrough in Cancer Chemotherapy
Mohd Imran Ansari, Chad R Johnson and Andrew Coop*
Department of Pharmaceutical Sciences, University of Maryland School of Pharmacy, USA
*Corresponding author: Andrew Coop, Department of Pharmaceutical Sciences, University of Maryland School of Pharmacy, Baltimore, USA
Submission: April 04, 2018;Published: May 15, 2018

ISSN: 2576-9170
Volume2 Issue1
Abstract
Among the 17 members of Poly (ADP-ribose) polymerase enzyme family, PARP-1 is a very crucial DNA repair enzyme and the level of PARP-1 is found elevated in several cancers such as ovarian, triple negative breast cancer. Inhibitors of PARP -1 is a new feature of anticancer drug development. In this mini-review, we covered the mode of action, recent development and approval of PARP-1 inhibitors. We also provide an insight on designing strategy of novel PARP-1 inhibitors and future direction.
Keywords: PARP inhibitors; Poly ADP ribose polymerase (PARP) family; Single strand break repair; Breast Cancer; Ovarian Cancer; DNA lipase
Abbreviations: PARP: Poly ADP Ribose Polymerase; SSBR: Single Strand Break Repair; CAT: C-Terminal Catalytic Domain; PARG: Poly(ADP-ribose) Glycohydrolase; SAR: Structure-Activity Relationship
Introduction
Among the 17 family members of poly ADP ribose polymerase (PARP) family, PARP-1 is comprehensively studied because of its critical roles in cancer [1]. The PARP family of proteins have been shown to regulate a vast number of cellular processes including DNA repair, stability of the genome (i.e. mammalian longevity), and programmed cell death.
The DNA repair function of PARP involves it’s assisting in the repair of single strand DNA “nicks” or breaks. The single strand break repair (SSBR) mechanism is activated when DNA damage occurs via exposure to DNA damaging agents both exogenous and endogenous (e.g. radiation exposure, chemotherapeutic agents). When PARP detects DNA damage it will bind to the damaged strand via N-terminal zinc fingers and undergoes a structural change. The synthesis of the polymeric adenosine diphosphate ribose (poly (ADP-ribose) or PAR) chain triggers the recruitment of other endogenous DNA repair enzymes [2]. However, DNA damage machinery, if Dysregulated, can lead to genetic mutations, which facilitate cancer development [3]. As certain cancer cell lines have been found to be defective in homologous recombination mechanisms, they may rely on PARP-mediated SSBR for survival. Therefore, inhibitors of PARP will prevent cancer cells from efficiently repairing their DNA, and increase their sensitivity to chemotherapeutics. For the last 3 decades, several PARP-1 inhibitors have been evaluated for their therapeutic potentials and later employed in clinical settings to treat cancer (e.g. breast cancer, ovarian cancer) [4].
PARP-1 was the first protein in the family identified and remains the best characterized. PARP-2 is most closely related to PARP-1, with 69% similarity in their catalytic domains. It was identified in PARP-1 deficient cells that still showed PARP activity [5]. While PARP-2 functions in a similar manner to PARP-1, it is much less abundant, contributing only 5-10% of total PARP activity [6]. It has been shown that PARP-1/PARP-2 oligomerizes when assisting in the repair of SSBR’s therefore PARP-1 inhibitors also inhibit PARP-2 [2]. PARP-1 inhibitors, in combination with anticancer agents such as DNA damaging chemotherapeutics have improved their efficiency [4]. PARP-1 inhibitors are also useful as stand-alone therapeutics in cancer types that have impaired DNA repair activities (e.g. breast cancer type 1/2 susceptibility protein (BRCA-1/2) mutations.
Mode of action
The PARP proteins are a family of stress response proteins approximately 116kDa that catalyze the transfer of ADP ribose to target proteins. Each of the 18 proteins is encoded by different genes, but all share a conserved catalytic domain. PARP is composed of four primary domains: the DNA binding domain, a caspase-cleaved domain, and auto-modification domain, and a catalytic domain. While it is appreciated that the PARP family is likely involved in several cellular processes including cell proliferation and death, the PARP family has gained notoriety for their involvement in DNA repair processes. Evidence for PARP-mediated DNA repair comes from the observation that the DNA damaging agents (e.g. chemotherapy and radiation) caused increased PARP activity [7].
PARP-1 and 2 are both activated by DNA SSBR’s. The DNA binding domain contains two zinc finger motifs which bind the damaged DNA and cause a conformational change bringing the N and C termini closer proximity. This action triggers the C-terminal catalytic domain (CAT) that hydrolyzes NAD+ to produce both linear and branched polymeric adenosine diphosphate ribose (PAR) polymers. This post-translational modification is known as PARylation, and it signals DNA ligase III, DNA polymerase beta, and other scaffolding proteins such as X-ray cross-complementing gene for DNA repair [8]. The PAR chain is subsequently degraded via poly (ADP-ribose) glycohydrolase (PARG) [9]. The oligomerization of PARP-1/2 has also been shown to stimulate PARP catalytic activity. This chain can extend up to 200 nucelotides before inducing any apoptotic processes via stimulation of the mitochondria to release apoptosis inducing factor (APF). Since NAD+ is required to generate these ADP-ribose monomers it has been thought that the synthesis of these chains depletes the cellular stores of NAD+. This is followed by progressive ATP depletion, which in turn induces necrotic cell death due to inhibition of glucose oxidation. More recently Andrabi et al. [10] showed that inhibition of hexakinase inhibits this activity [9]. PARP is inactivated by caspase-3 cleavage which proteolytically cleaves PARP into 89kDA and 24kDA fragments-a common early indicator of apoptosis.
Small molecule PARP inhibitors have been extensively studied as a new class of anticancer agents. Each is an analogue of nicotinamide, and several potent and selective analogues have been reported thus far. Although PARP inhibitors have been studied over 30 years, the exact mechanism of their actions is still not fully understood. However, a general consensus appears to agree on two possible modes of their actions:
- PARP-1 inhibitors interact with and inhibit PARP-1 by trapping the DNA-PARP-1 complex-hence blocking the DNA repair activities (e.g. inhibition of PARylation) and causing radio/chemopotentiation.
- Some PARP inhibitors may also trap PARP-1/2 DNA complex by way of an allosteric effect as shown by Murai [4,5,10,11].
Recent advancement and SAR of PARP inhibitors
The PARP-1 catalytic domain has been successfully crystallized without an exogenous ligand and co-crystallized with NAD+, which has facilitated the rational design of PARP-1 inhibitors. PARP-1 inhibitors serve as competitive catalytic inhibitors with NAD+, which block the auto-PARylation post-DNA binding-an important step in successfully mounting a DNA repair response. The early knowledge and research on the basic pharmacophore nicotinamide, which is a weak PARP inhibitor, derived an overlay to design and develop next-generation PARP inhibitors (Figure 1) [11].
figure 1: Required feature for PARP-1 inhibitor.

figure 2: Clinically important PARP-1 inhibitors.
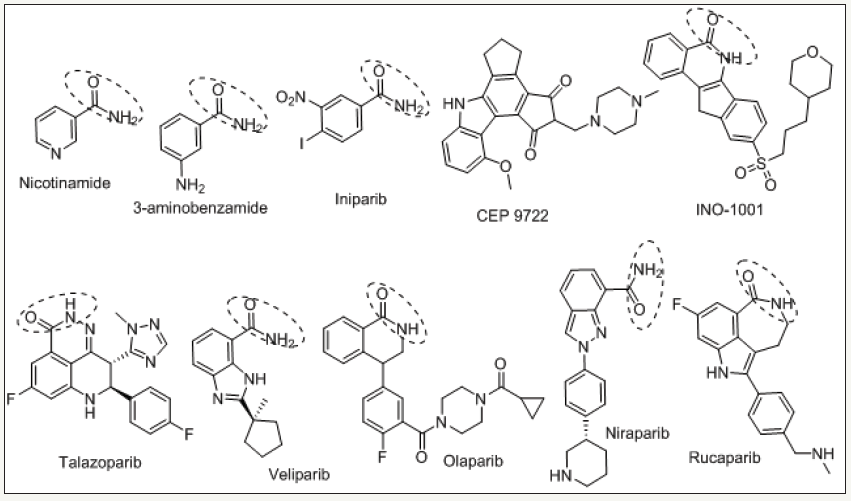
Iniparib (Figure 2), a PARP inhibitor developed against triple negative breast cancer, failed in phase III clinical trials [12]. In recent years, researchers have made an effort on designing a library of small molecule PARP inhibitors that offered improved potency to first generation drugs such as Iniparib. This helped pave the way for the next-generation of PARP inhibitors (Figure 2) [13]. From SAR studies of screened compounds against PARP, it was proposed and validated that restricting the degree of conformational freedom of the amide bond via formation of a bicycle or pseudo-ring system could enhance the PARP inhibitors performance and potency (Figure 1). To support this hypothesis several hetero cycles has been developed and all these molecules were found potent PARP inhibitors. This work has helped in the development of clinically relevant PARP inhibitors [14].
Griffin [19] designed and developed benzoxazole carboxamides and imidazoles derivatives with Veliparib being the most potent compound of that series (Figure 1). Veliparib’s improved potency was attributed to the pseudo-intra molecular hydrogen bond between the imidazole nitrogen and the amidic NH that restricted the degree of freedom of amide [15]. Ruf and co-workers rationally designed several PARP inhibitors based on the co-crystal structures of PARP inhibitors with the PARP-1 protein [16]. They concluded that hydrogen bonding between PARP-1 amino acid residues and amide core of PARP-1 inhibitors was a useful feature in the design of potent inhibitors [17]. Based upon these and other structure activity studies, key structural features have been identified in the design of potent PARP-1 inhibitors such as:
- A conformationally restricted amide moiety attributed to pseudo-intra molecular hydrogen bonding, as stated above- (1 & 2 in Figure 1).
- Bulky group decoration on the core scaffold improves drug potency represented by Olaparib and Rucaparib (3 in Figure 1).
- Aryl group functionality (4 in Figure 1).
For some PARP-1 inhibitors, such as talazoparib, stereochemistry is important for activity. One of the talazoparib isomers was shown to exhibit 300 times improved potency over the other (Figure 2) [18,19].
FDA approval
FDA-approved PARP-1 inhibitors, such as Olaparib, Rucaparib, and Niraparib are successful drugs. Olaparib was produced by the Astra Zeneca as the first PARP-1 inhibitor for ovarian cancer. Olaparib was approved to be used as mono-therapy in ovarian cancer cases that have germline BRCA mutations. While not showing therapeutic potential against estrogen-positive breast cancer, it was approved for germline BRCA HER2-negative breast cancer and for patients that are tested with adjuvant, neoadjuvant. Another FDA-approved single-agent PARP inhibitor against ovarian cancer, Rucaparib, has been employed in patients that have either somatic or germilne BRCA mutation and the patients that have gone through minimum of two chemotherapeutics regimens before that. Niraparib, an orally active PARP inhibitor, has been successfully approved in patients with recurrent ovarian, fallopian tube, or primary peritoneal cancer that have had either complete or partial response to platinum-based chemotherapy [20,21].
Conclusion and Future Prospective
For over three decades, medicinal chemists have focused on designing, characterizing, and validating PARP-1 inhibitors for their potency and activity against cancer. In addition, several groups have worked on comprehensively characterizing the mechanistic aspects of PARP activation and how inhibitors modulate their function. Current PARP-1 inhibitors on the market are non-selective and inhibit the function of both PARP-1 and PARP-2 proteins due to the oligermization of PARP-1/2 and their high structural homology. Further efforts in characterizing their roles in DNA SSBR repair and the rational design of selective moieties will prove to be an effective method to develop more potent inhibitors. Additionally, studying the roles and biological implications of other PARP family proteins will open new drug discovery and therapeutic avenues.
References
- Hottiger MO, Hassa PO, Lüscher B, Schüler H, Koch Nolte F (2010) Toward a unified nomenclature for mammalian ADP ribosyl transferases. Trends Biochem Sci 35(4): 208-219.
- Miwa M, Masutani M (2007) Poly ADP ribosylation and cancer. Cancer Sci 98(10): 1528-1535.
- Plummer R (2010) Perspective on the pipeline of drugs being developed with modulation of DNA damage as a target. Clin Cancer Res 16(18): 4527-4531.
- Ferraris DV (2010) Evolution of Poly (ADP-ribose) Polymerase-1 (PARP- 1) Inhibitors. From Concept to Clinic J Med Chem 53(12): 4561-4584.
- Liscio P, Camaioni E, Carotti A, Pellicciari R, Macchiarulo A (2013) From Polypharmacology to Target Specificity: The Case of PARP Inhibitors. Curr Top Med Chem 13(23): 2939-2954.
- Amé JC, Spenlehauer C, de Murcia G (2004) The PARP super family. BioEssays 26(8): 882-893.
- Cerrato A, Morra F, Celetti A (2016) Use of poly ADP ribose polymerase [PARP] inhibitors in cancer cells bearing DDR defects: the rationale for their inclusion in the clinic. J Exp Clin Cancer Res 35: 179.
- Skidmore CJ, Davies MI, Goodwin PM, Halldorsson H, Lewis PJ, et al. (1979) The involvement of poly(ADP-ribose) polymerase in the degradation of NAD caused by gamma radiation and N-methyl-Nnitrosourea. Eur J Biochem 101(1): 135-142.
- Isabelle M, Moreel X, Gagné JP, Rouleau M, Ethier C, et al. (2010) Investigation of PARP-1 PARP-2 and PARG interactomes by affinity purification mass spectrometry. Proteome Sci 8: 22.
- Andrabi, SA, Umanaha GKE, Stevensa CDA, Karuppagoundera SS, Gagnée JP, et al. (2014) Poly (ADP-ribose) polymerase dependent energy depletion occurs through inhibition of glycolysis. Proc Natl Acad Sci 111(28):10209-10214.
- Murai J, Huang SN, Das BB, Renaud A, Zhang Y, et al. (2012) Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res 72(21): 5588- 5599.
- Banasik M, Komura H, Shimoyama M, Ueda K (1992) Specific inhibitors of poly (ADP-ribose) synthetase and mono (ADP ribosyl) transferase. J Biol Chem 267(3): 1569-1575.
- Shaughnessy J, Schwartzberg L, Danso MA, Miller KD, Rugo HS, et al. (2014) Phase III Study of Iniparib Plus Gemcitabine and Carboplatin versus Gemcitabine and Carboplatin in Patients With Metastatic Triple Negative Breast Cancer. J Clin Oncol 32(34): 3840-3847.
- Threadgill MD (2015) 5-Aminoisoquinolin-1-one (5-AIQ), a watersoluble inhibitor of the poly(ADP-ribose)polymerases (PARPs). Curr Med Chem 22(33): 3807-3829.
- Baptista SJ, Silva MMC, Moroni E, Meli M, Colombo G, et al. (2017) Novel PARP-1 inhibitor scaffolds disclosed by a dynamic structure based pharmacophore approach. PLoS ONE 12(1): e0170846.
- Ferraris D, Ko YS, Pahutski T, Ficco RP Serdyuk L, et al. (2003) Design and synthesis of poly ADP ribose polymerase 1 inhibitors. Biological evaluation of aza-5[H] phenanthridin-6-ones as potent, aqueous soluble compounds for the treatment of ischemic injuries. J Med Chem 46(14): 3138-3151.
- Shinkwin AE, Whish WJ, Threadgill MD (1999) Synthesis of thiophene carboxamides, thieno[3,4-c]pyridin-4(5H)-ones and thieno[3,4-d] pyrimidin-4(3H)-ones and preliminary evaluation as inhibitors of poly(ADP-ribose)polymerase (PARP). Bioorg Med Chem 7(2): 297-308.
- Penning TD, Zhu GD, Gandhi VB, Gong J, Liu X, et al. (2009) Discovery of the Poly(ADP-ribose) polymerase (PARP) inhibitor 2-[(R)-2- methylpyrrolidin-2-yl]-1H-benzimidazole-4-carboxamide (ABT-888) for the treatment of cancer. J Med Chem 52(2): 514-523.
- White AW, Almassy R, Calvert AH, Curtin NJ, Griffin RJ, et al. (2000) Synthesis and biological properties of benzimidazole inhibitors of the DNA repair enzyme poly (ADP-ribose) polymerase. J Med Chem 43(22): 4084-4097.
- Ruf A, de Murcia G, Schulz GE (1998) Inhibitor and NAD+ binding to poly (ADP-ribose) polymerase as derived from crystal structures and homology modeling. Biochemistry 37(11): 3893-3900.
- Wang B, Chu D, Feng Y, Shen Y, Aoyagi-Scharber M, et al. (2016) Discovery And Characterization Of (8S,9R)-5-Fluoro-8-(4-Fluorophenyl)-9-(1- Methyl-1H-1,2,4-Triazol-5-Yl)-2,7,8,9-Tetrahydro-3H-Pyrido[4,3,2-De] Phthalazin-3-One (BMN673,alazoparib), A Novel Highly Potent And Orally Efficacious Poly(ADP-Ribose)Polymerase-1/2 Inhibitor As An Anticancer Agent. J Med Chem 59(1): 335-357.
- https://www.fda.gov/Drugs/InformationOnDrugs/ucm412998.htm
© 2018 Andrew Coop. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






