- Submissions

Full Text
Modern Approaches in Drug Designing
Importance of Computer Aided Screening Techniques in Drug Design: An Editorial
Shriram H Bairagi*
Department of Drug Design, Ideal College of Pharmacy and Research, India
*Corresponding author: Shriram H Bairagi, Department of Drug Design, Ideal College of Pharmacy and Research, Kalyan, 421306,India
Submission: February 8, 2018;Published: March 28, 2018

ISSN: 2576-9170
Volume1 Issue5
Introduction
Drug design and development is a very complicated, time consuming process and there are many factors responsible for the failure of different drugs such as lack of effectiveness, side effects, poor pharmacokinetics, and marketable reasons. “Drug discovery” has historically been a stronghold of chemistry, especially natural products chemistry and synthetic organic chemistry. Chemistry is thus inherently playing a major impact on drug discovery, since other disciplines cannot create novel small molecules. In other words, “drug discovery and development” cannot happen without chemists.
Molecular target-based drug discovery with continuous improvement in the quality and diversity of compound libraries has been making steady progress. “Structural biology”, “computational biology”, “chemical biology”, and “chemical genetics” are now fully incorporated into modern drug discovery through target- based approaches. With these modern tools in hand, rational drug design, exploiting combinatorial chemistry for focused libraries, becomes possible. Designing hybrid molecules having a dual mode of action is a good example and provides a promising approach to modern drug discovery. Natural products chemistry provides sufficiently sophisticated lead structures for drug discovery. A combination of natural products chemistry and focused library synthesis furnishes a powerful approach to drug discovery. The terms “biology-oriented synthesis (BIOS)” and “function-oriented synthesis (FOS)” have recently emerged as a logical evolution in “chemical genetics” for exploring the intrinsic complexity and diversity of the structures of natural products. Moreover, the use of combinatorial biosynthesis, taking advantage of particular gene clusters as a tool for creating a library of highly sophisticated natural product leads, provides another fascinating approach to modern drug discovery [1].
The chemical structural space using an ingenious computer program called Scaffold Hunter, this generates maps of a selected chemical structural space based on structural criteria and enables the researchers to retrieve possible molecules and approaches towards biological activity on the computer screen. The Scaffold Hunter searches for structural moieties that resemble already known structures with a particular biological characteristic so they use these as a basis and experiment with different chemical information to synthesis new compounds [2].
We have seen a technological switch in the methodology used by drug discovery team; the serendipitous, individual approach that you once used has been largely replaced by rational drug design using a multidisciplinary, team approach which uses new computeraided drug design methods that speed up the drug discovery process and generate more accurate, viable lead compounds. In turn, your job description and skill requirements have changed dramatically. No longer do you mainly concern yourself with synthetic problems, but the increased use of computer-aided drug design has created greater opportunities for you to work with experts from a range of specialties and for you to understand and predict the impact of your daily work on the drug design process as a whole.
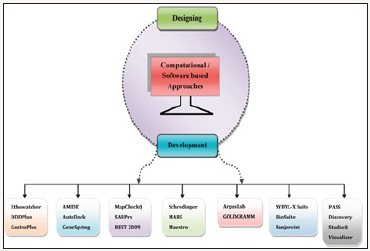
The molecular docking is a technique used to study orientation of one molecule to a second, when bound to each other to form a stable complex [3]. Molecular docking predicts ligand binding to its receptor or target protein. Molecular docking is used to recognize and optimize drugs by examining and modelling molecular interactions between ligand and target macromolecules. Molecular docking is used to generate multiple ligand conformations and orientations and the most appropriate ones are selected [4,5]. The several molecular docking tools are ArgusDock, Schrodinger, DOCK, BioSuite, FRED, GRAMM, eHITS, Maestro, AutoDock, SYBYL-X Suite and FTDock. Molecular modelling involves scoring methods that are used to rank the affinity of ligands to bind to the active site of a receptor. In virtual screening, compounds are docked and then scored to find out the one which is more likely to bind tightly to the target macromolecule [6].
The 3D pharmacophore search is an imperative, vigorous and simple method to quickly recognize lead compounds towards a preferred target. Pharmacophore is a specific 3D arrangement of functional groups within a molecular framework that are essential to attach to an active site of an enzyme or bind to a macromolecule. Once a pharmacophore is defined, the medicinal chemist utilizes the 3D database search tools to find novel compounds that are suitable for the pharmacophore model. The modern drug design process has been used to make it one of the most successful computational tools because it efficiently identify and optimize lead focus combinatorial libraries and helps in virtual high-throughput screening [7]. The novel and protracted techniques of QSAR analysis have advanced to transform pharmacophore detail into QSAR models that, in turn can be used as virtual high-throughput screens for activity profiling of a library [8]. An additional broad idea of descriptor pharmacophore was presented, which uses a variable selection QSAR as a division of molecular descriptors that afford the maximum statistically important structure-activity relationship. These approaches comprise of partial least squares and K-nearest neighbours. Thus, chemical similarity searches involving descriptor pharmacophores produces resourceful pulling out of chemical databases or virtual libraries to find out compounds with anticipated biological activities [9] (Figure 1 & 2).
Figure 1:

Figure 2:
References
- Iwao Ojima (2008) Modern molecular approaches to drug design and discovery. Acc Chem Res 41(1): 2-3.
- Herbert Waldmann (2012) Biology-oriented synthesis (BIOS)-cancer drugs based on natural models. Max Planck Institute of Molecular Physiology, Dortmund.
- Soloman AK (2008) Molecular modelling and drug design. MB publisher, Chennai, India.
- Lesk AJM (2002) Introduction to bioinformatics. Oxford University press inc., New York, USA.
- Perdo HL (2010) A systematic review on CADD: Docking and Scoring. JMPI: pp. 47-51.
- http:en.wikipedia.org/wiki/
- Meyer EF, Swanson SM, Williams JA (2000) Molecular modeling and drug design. Pharmacol Ther 85(3): 113-121.
- Hopfinger AJ, Duca JS (2000) Extraction of pharmacophore information from high-throughput Screens. Curr Opin Bio technol 11(1): 97-103.
- Bleicher KH, Bohm HJ, Muller K, Alanine AI (2003) Hit and lead generation: Beyond high-throughput screening. Nat Rev Drug Discov 2(5): 369-378.
© 2018 Shriram H Bairagi. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






