- Submissions

Full Text
Modern Approaches in Drug Designing
Computer Aided Drug Designing of Novel Inhibitors against Pfama-1
Miti Shah, Nilanjan Roy and Pinakin Dhandhukia*
Department of Research in Biotechnology and Allied Sciences, Ashok and Rita Patel Institute of Integrated Study, India
*Corresponding author: Pinakin Dhandhukia, Ashok and Rita Patel Institute of Integrated Study and Research in Biotechnology and Allied Sciences (ARIBAS), New Vidhyanagar -388121, Gujarat, India
Submission: August 10, 2017; Published: January 03, 2018

ISSN : 2576-9170Volume1 Issue4
Abstract
The invasion of Plasmodium in RBC is the most important pathogenic step. Further proliferation is stopped if any drug or antibody can inhibit at this stage. Among two major invasive surface proteins PfRH5 and AMA-1, only multiple AMA-1 structures viz. 3ZWZ, 2Q8B, 2Z8V, 3SRJ are available at time of study which was selected to obtain information about the common ligand-bound interface residues. Three active sites were created and the amino acid properties provided information that some of these are polymorphic and some are hydrophobic and conserved. Docking was done using the ZINC library on the Web server. Interactions of H-bond of the crystal structure and docking results revealed that ASN223 and ILE225 amino acids were the common H-linkage formation in all structure. ILE225 is a polymorph that cannot be considered as an interaction, whereas ASN223 is a very important polar interacting residue. However, from the docking results, only the compounds interacting with ASN223 were analyzed. ZINC80342995 and ZINC95042198 have been selected as active compounds. Both have properties in the range of the ideal drug according to ADME/T properties. However, in vitro analysis will be needed to analyze the actual potential of these candidates to prevent invasion.
Keywords: Malaria; Drug design; Invasion inhibition
Introduction
Human malaria caused by the five Plasmodium sp. viz. P falciparum, P. vivax, P. malariae, P. ovale and P. knowlesi, is transmitted by the female anopheles mosquitoes, and remains the leading cause of death worldwide [1]. The causative agent of malaria is Plasmodium parasite, which completes its life cycle in the two hosts, mosquitos and humans [2]. Current anti-malarial drugs include chloroquine, primaquine, quinine, quinoline, folate inhibitor, artimisin in and their derivatives are employed for the treatment of malaria. Combination therapy based on artimisin in is a widely used treatment. It is combined with long term effective drug for better cure [3]. Different drugs work at different stage in the life cycle [4,5]. However, continuous exposure to the drugs make parasite resistant and eventually it become predominant [68]. This requires the design of new drugs that should be effective, provide the medicine at a reasonable time, less toxic and less expensive [9]. Researchers are developing several antibodies but, due to polymorphic nature of parasite surface proteins, it remained ineffective [10].
The parasite life cycle is a complex process involving sexual and asexual reproduction, which is complemented by two hosts, mosquitoes and humans. Asexual reproduction of a parasite of malaria present in humans that contains hepatic and erythrocytic conditions, while sexual reproduction is completed in mosquitoes [11]. During the erythrocytic phase, the parasitic invasion of erythrocytes is the most critical and pathogenic step. This is a multistage process involving a cascade of molecular events between merozoites and host erythrocytes [12]. Malaria parasite surface proteins act as a ligand, which is one of the two molecular protein families known as EBL (erythrocyte binding) and RBL (reticulocyte binding) proteins. This EBL family of protein is also known as the duffy binding (DBL) protein family. In P. falciparum, members of these families are termed PfEBL (P. falciparum erythrocyte bindinglike) and PfRH (P. falciparum reticulocyte binding-like homolog) proteins [13,14].
Apart from these two protein families (involve in secondary interaction), there are other proteins involved in invasion process e.g. Glycosyl Phosphatidyl Inositol (GPI) anchored membrane proteins like Merozoite Surface Proteins (MSPs), Apical Membrane Antigen-1 (AMA-1), motor associated protein (TRAP) etc. [1].
AMA-1, type-1 integral membrane protein, is refractory to gene deletion and highly conserved throughout the phylum [1,15]. Among 622 amino acids, 10% are polymorphic in nature [16]. From all the 3 domains, Domain I and domain II are highly conserved throughout the Apicomplexa while domain III is less conserved. Same way the polymorphism found more in Domain I and lesser in domain III. Domain I contains several loops which are Ia-If. Hydrophobic conserved cleft (region) of domain I is surrounded by the polymorphic residues [17,18]. Domain I and II share a common core topology called as PAN domain or Apple fold consisting five- strand (3 sheet flanked by single a helix at one side and three stranded sheet at other side [19]. AMA-1 form complex with RON i.e. Rhoptry Neck Protein releases from the Rhoptry secretary organelle present in parasite [15]. It is inserted into the host plasma membrane (erythrocyte membrane) and form a complex of RON2/4/5/8 [20]. RON protein family of parasite is highly conserved. This complex contains RON2 (assumed to contain 3 hydrophobic helices), RON5 (contain only one predicted hydrophobic helix) and RON4, RON8 (both appear to be soluble proteins) [21].
In present study, AMA-1 structures viz., 3ZWZ, 2Q8B, 2Z8V, 3SRJ were selected to obtain the information of common interface residues. Using these residues, three active sites were generated. Properties of amino acids gave the information that some of amino acids are polymorphic and some are conserved hydrophobic. Docking was carried out with a ZINC library on a web server. H-bond interactions of crystal structure and docking results were analyzed.
Materials and Methods
Retrieval and study of AMA-1 crystal structure
AMA-1 crystal structures bound with different peptides were retrieved from RCSB PDB (http://www.rcsb.org/). PDB ID of selected structures were 3SRJ, 3ZWZ, 2Z8V, and 2Q8B bound with inhibitory peptide R1, small peptide of RON2 erythrocyte receptor, IgNAR antibody and growth inhibitory antibody, respectively [19,22,23]. Protein structure and their amino acid interactions were analyzed by Discovery Studio Visualizer 3.0 (Accelrys Inc., San Diego, CA, USA) [24]. Rama chandran plot of all crystal structures Table 1: Different parameters of active sites.were generated using RAMPAGE (University of Cambridge, Dept. of Biochemistry).
Selection of common interface residues
All bound structures contain interface residues which were found by Inter Pro Surf server (http://curie.utmb.edu/prosurf. html/) [25]. A, A, A and B, A chain of AMA-1 from 2Q8B, 2Z8V, 3SRJ and 3ZWZ were submitted for analysis of interface residues. All resulted residues of four structures were compared and common amino acids were selected using Discovery Studio Visualizer 3.0 (http://accelrys.com/).
Active site generation from interface residues
These selected residues gave the buried pockets where all the peptides were interacting. Active site residues were covering larger area so it was split into three small active sites covering different areas.
Docking studies
Generated active sites were used further for docking. Different parameters of active site were set according to auto dock vina (Table 1). For that AMA-1 from 3ZWZ PDB structure was selected. Docking was done by idock, a multithreaded virtual screening tool (http://istar.cse.cuhk.edu.hk/idock) which is linked with the ZINC databases [26]. Filtration of ZINC database were done using Lipinski's rule of five and other parameters were set according to ideal characteristics of drugs which act on blood stages (Table 2). The results obtained after docking were visualized in an iview (interactive WebGL visualize for protein-ligand complex) [27].
Table 1: Different parameters of active sites.

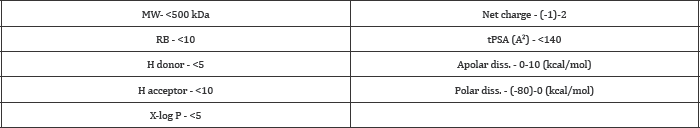
Table 2: Filtrations applied for ZINC database.
Selection of hits
Hits were selected on the bases of their interaction with specific hydrophobic, conserved, non polymorphic amino acids from the given residues and forming maximum bonds. For that, residues characteristics were studied from literatures. Comparison of H-bond formation in four crystal structures and docking result was done which were also used in selection on specific amino acids.
ADME/T properties prediction
ADME/T predictions of selected compounds were carried out using admet SAR (http://admetexp.org/) which is a user friendly interface [28]. Compounds, falling in acceptable range, were selected.
Results and Discussion
Study of AMA-1 crystal structure
Among the four crystal structures of AMA-1, Ramachandran plot of 3ZWZ was created which is shown in Figure 1. It shows 0% amino acids are present in outer region of a plot while 2% are present in allowed region. Remaining residues are present in favored region. Interface residues of different structures obtained from Inter Pro Surf tool [25] were listed in excel sheet for comparison. About 12 common interacting residues present in all the structures are shown in Table 3 with its position number. Some structures are also having two chains of AMA-1 and one or two interacting amino acid might change but both were considered. This information will further useful for analysis of binding pattern. Individual crystal structures are also having numbers of other interacting amino acids [29] other than listed in Table 3. These common residues (labeled in blue color) were shown in Figure 2. Different residues were forming loops, helix, turns and sheets (Table 3 and Figure 1,2).
Table 3: List of common interface residues.
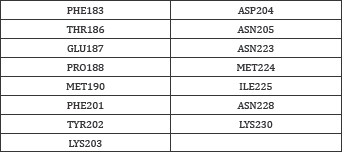
Figure 1: AMA-1 structure (a) its Ramachandran plot (b) generated using (RAMPAGE).
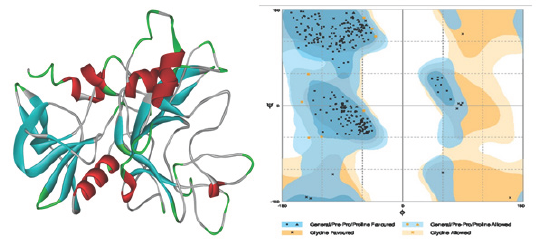
Figure 2: Common interface residues (blue labeled) in AMA-1 crystal structure.

Active site generation and selection
Interface residues, act as an active sites [30], were split into 3 different active sites. Among all these residual active site, 1st active site which contains PHE183, THR186, GLU187, PRO188, MET190 amino acids were forming the loop. It was proven that loop may Interface residues, act as an active sites [30], were split into 3 change the shape and their positions as well as they do not play a different active sites. Among all these residual active site, 1st active role in protein stability [31]. There might be possibility that same compound binds with one or more active residues. Other two active sites were also having the loop forming amino acids but they even contain amino acids which were involve in helix, turns and sheets surrounding it. These sites were nearer to each other so there might be possibility that compounds interacting with 2nd active site may also interact with amino acids present in 3rd active site. Therefore it should be determined that these interactions are favorable of not.
Docking results and ADME/T studies
The docking result, obtained from idock, was saved in a excel sheet which contain the information required for binding analysis. idock gave 17,746,988 numbers of compounds from the ZINC database after filtrations. If the library generated in idock exceed 1,00,000 compound limit than it will take randomized 1,00,000 compound from that library. So, here docking was carried out with randomly selected 1,00,000 compounds.
The hydrogen bonding and their interactions was visualized in iview [27]. The hydrogen bonds forming amino acids of crystal structures with the peptides were shown in Figure 3 where hydrogen bonds highlighted in green colored dotted line. This analysis gave the information about amino acids and exact atoms of amino acids which play a role in H-bond formation [32].
Similar analysis was performed for all docking results. The top 20 interacting compounds were taken in consideration and their interacting amino acid: atom information was combined in Active Site (log) 1, 2, 3 in a excel sheet. Same information of crystal structures were also taken from Figure 3 in excel sheet. Here, Table 4 is showing information of amino acids participating in the hydrogen bond formation and their position in known structure of AMA-1. If any similar amino acids or atoms interact in every structure and active site, they were highlighted in same color (Table 4 and Figure 3).
Table 4: H-bond forming amino acids of crystal structures and docking results.
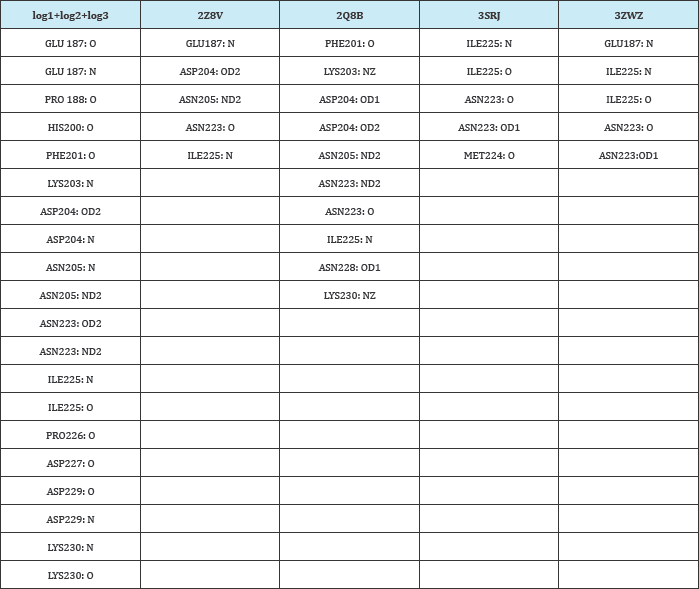
Figure 3: H-bond interaction of selected amino acids of AMA-1 with peptides and antibodies. (A)3SRJ (B)3ZWZ (C)2Z8V (D)2Q8B. Only interactive residues were shown and labeled in red color.
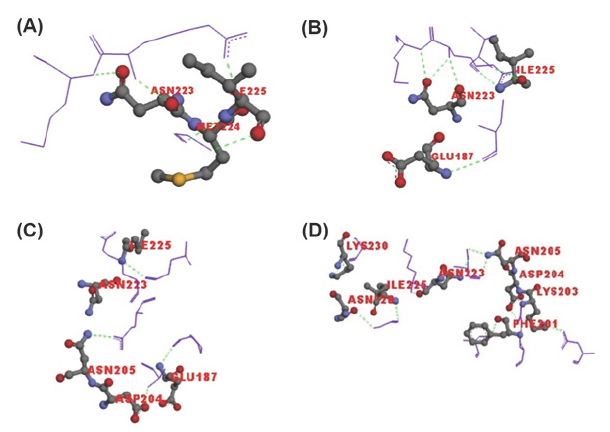
From this interaction analysis, two amino acids were found commonly interacting in four structures and reciprocated in docking results. ASN223 (OD1, ND2- are oxygen and nitrogen present at delta position respectively) and ILE225 (N) amino acids were forming H-bonds with the peptides and docked compounds. Apart from these, other amino acids were also forming H-bonds. From the literature review the properties of different amino acids play role in the interaction is summarized in (Table 5).
Table 5: Properties of selected amino acids.
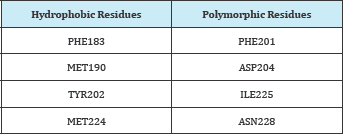
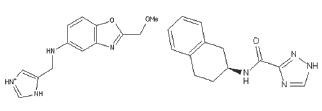
It was proven that ILE225 is polymorphic and due to this nature it cannot be considered further as mutation at this residue abrogate the binding with peptides Coley et al. [19]. Second amino acid ASN223 reported in making very important polar interaction [22]. Though it does not fall in a hydrophobic conserved amino acids however does not change during every cell cycle. Compounds which interact with ANS223 that also interact with side amino acids and block that groove so that RBC receptor or other peptides cannot bind with it. Therefore compounds having top docking scores and specifically interacting with ASN223 amino acid were selected. Two compounds ZINC80342995 and ZINC95042198 were predicted as good effective hits. Structures of these compounds are shown in Figure 4 and their interactions in docking with AMA-1 are shown in (Figure 4,5).
Figure 4: ZINC80342995 (A) and ZINC95042198 (B).
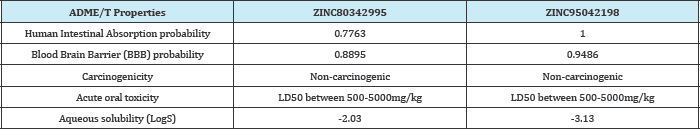
These two compounds were interacting with ASN223 and other amino acids. All the H-bonds are shown in sky blue color. More H-bonds form more stronger interaction and it also require more energy for removal of bound compound from the active site. Therefore it will be favorable if any compound interacts with more H-bonds. Here, ZINC80342995 and ZINC95042198 are forming 9 and 4 H-bonds respectively. Therefore, ZINC80342995 could be a better inhibitor. Both the structures are satisfying the Lipinski's rule of five and also the polar and non-polar dissovation, tPSA (topological polar surface area) range. The ADME/T properties of both compounds are given in Table 6 which shows that it is not harmful for human consumption. Therefore, we can conclude that both compounds are having ideal drug properties. But all the results were predicted by the software which further needed to confirm by in vitro and then in vivo experiments to prove its activity (Table 6).
Figure 5: ZINC80342995 and ZINC95042198 docking interaction (H-bonds) with AMA-1.
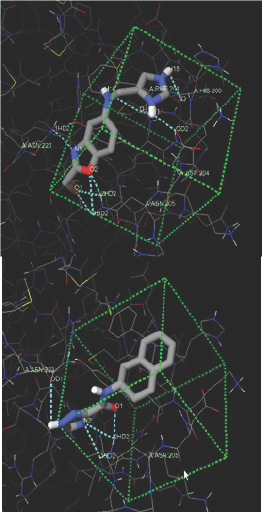
Table 6: ADME/T properties of two compounds with values.
References
- Cowman AF, Crabb BS (2006) Invasion of red blood cells by malaria parasites. Cell 124(4): 755-766.
- Kakkilaya BS (2006) Malaria Site: All about malaria.
- Biamonte MA, Wanner J, Le Roch KG (2013) Recent advances in malaria
- drug discovery. Bioorg Med Chem Lett 23(10): 2829-2843.
- Bruce-Chwatt LJ (1962) Classification of anti-malarial drugs in relation to different stages in the life-cycle of the parasite: commentary on a diagram. Bull World Health Organ 27:287-290.
- Peters W (1973) Anti-malarial drugs and their actions. Postgrad Med J 49(574): 573-583.
- Krishna S, Uhlemann AC, Haynes RK (2004) Artemisinins: mechanisms of action and potential for resistance. Drug Resist Updat 7(4-5): 233-244.
- Hoffman SL (1986) Treatment of Malaria. Cincs Trop Med Commun Dis 1: 171-224.
- WHO (2010) guideline for the treatment of malaria. (2nd edn), Guidel treatmalar 2010: 210.
- Fidock DA, Rosenthal PJ, Croft SL, Brun R, Nwaka S (2004) Anti-malarial drug discovery: efficacy models for compound screening. Nat Rev Drug Discov 3(6): 509-520.
- Cowan GJM, Creasey AM, Dhanasarnsombut K, Thomas AW, Remarque EJ, et al. (2011) A malaria vaccine based on the polymorphic block 2 region of MSP-1 that elicits a broad serotype-spanning immune response. PLoS One 6(10): e26616.
- Cowman AF, Berry D, Baum J (2012) The cellular and molecular basis for malaria parasite invasion of the human red blood cell. J Cell Biol 198(6): 961-971.
- 12. Elnahas NS (2012) Molecular mechanisms of malarial parasite invasion. PUJ 5:85-92.
- Wellems TE, Fairhurst RM (2012) An evolving picture of the interactions between malaria parasites and their host erythrocytes. Cell Res 22(3): 453-456.
- Tham WH, Healer J, Cowman AF (2012) Erythrocyte and reticulocyte binding-like proteins of P. falciparum. Trends Parasitol 28(1): 23-30.
- Cao J, Kaneko O, Thongkukiatkul A, Tachibana M, Otsuki H, et al. (2009) Rhoptry neck protein RON2 forms a complex with microneme protein AMA1 in P.falciparum merozoites. Parasitol Int 58(1): 29-35.
- Healer J, Triglia T, Hodder AN, Gemmill AW, Cowman AF (2005) Functional analysis of P. falciparum apical membrane antigen 1 utilizing interspecies domains. Infect Immun 73(4): 2444-2451.
- Macraild CA, Anders RF, Foley M, Norton RS (2011) Apical Membrane Antigen 1 as an Anti-Malarial Drug Target. Curr Top Med Chem 11(16): 2039-2047.
- Cortes A, Mellombo M, Mueller I, Benet A, Reeder JC, et al. (2003) Geographical Structure of Diversity and Differences between Symptomatic and Asymptomatic Infections for P falciparum Vaccine Candidate AMA1. Infect Immun 71(3): 1416-1426.
- Coley AM, Gupta A, Murphy VJ, Bai T, Kim H, et al. (2007) Structure of the malaria antigen AMA1 in complex with a growth-inhibitory antibody PLoS Pathog 3(9): 1308-1319.
- Lamarque M, Besteiro S, Papoin J, Roques M, Vulliez-Le Normand B, et al. (2011) The RON2-AMA1 interaction is a critical step in moving junction-dependent invasion by apicomplexan parasites. PLoS Pathog 7(2): e1001276.
- Srinivasan P, Beatty WL, Diouf A, Herrera R, Ambroggio X, et al. (2011) Binding of Plasmodium merozoite proteins RON2 and AMA1 triggers commitment to invasion. Proc Natl Acad Sci USA 108(32): 13275-13280.
- Normand BVL, Tonkin M, Lamarque MH, Langer S, Hoos S, et al. (2012) Structural and functional insights into the malaria parasite moving junction complex. PLoS Pathog 8(6): e1002755.
- Henderson KA, Streltsov VA, Coley AM, Dolezal O, Hudson PJ, et al. (2007) Structure of an IgNAR-AMA1 complex: targeting a conserved hydrophobic cleft broadens malarial strain recognition. Structure 15(11): 1452-1466.
- Arnott A, Mueller I, Ramsland PA, Siba PM, Reeder JC, et al. (2013) Global Population Structure of the Genes Encoding the Malaria Vaccine Candidate , Plasmodium vivax Apical Membrane Antigen 1 ( PvAMA1 ). PLoS Negl Trop Dis 7(10): e2506.
- Negi SS, Schein CH, Oezguen N, Power TD, Braun W, et al. (2007) InterProSurf: A web server for predicting interacting sites on protein surfaces. Bioinformatics 23(24): 3397-3399.
- Li H, Leung KS, Wong MH (2012) idock: A multi threaded virtual screening tool for flexible ligand docking. IEEE, California, USA, pp. 7784.
- Li H, Leung K, Nakane T, Wong M (2014) iview : an interactive Web GL visualizer for protein-ligand complex. BMC Bioinformatics 15: 56.
- Cheng F, Li W, Zhou Y, Shen J, Zengrui W, et al. (2012) admetSAR: a comprehensive source and free tool for assessment of chemical ADMET properties. J Chem Inf Model 52(11): 3099-3105.
- 29. Kortagere S, Cocklin S, Loraiya K, et al (2010) Structure based design of novel small molecule inhibitors to apical membrane antigen-1 of P falciparum. Intell Syst Mol Biol 2010: 1.
- Plouffe D, Brinker A, McNamara C, Henson K, Kato N, et al. (2008) In silico activity profiling reveals the mechanism of action of antimalarials discovered in a high-throughput screen. Proc Natl Acad Sci USA 105(26): 9059-9064.
- Fetrow JS (1995) Omega loos: nonregular secondary structures significant in protein function and stability. FASEB J 9(9): 708-717.
- Chaudhary KK, Prasad CVSS (2014) Virtual Screening of compounds to 1-deoxy-D- xylulose 5-phosphate reductoisomerase ( DXR ) from P. falciparum. Bioinformation 10(6): 358-364.
© 2018 Miti Shah, et al. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






