- Submissions

Full Text
Journal of Biotechnology & Bioresearch
Analysis of the vcgC Gene in Vibrio vulnificus Isolates from the Texas Coastal Bend region of the Gulf of Mexico
Gregory W Buck1*, LarReshia I Brumfield1, Stephanie Dudics Giagnocavo1,2, Danielle S Perkins1,3, Alvaro Ortola Tortosa1, Tolulope B Okuyemi1, Joshua S Carbaugh1, Githzette M Planas Costas1 and Gabriel D Ramirez1
1Department of Life Sciences, Texas A&M University-Corpus Christi, Corpus Christi, Texas, USA
2Thiel College, Greenville, Pennsylvania, USA
3Washburn University, Topeka, Kansas, USA
*Corresponding author:Gregory W Buck, Department of Life Sciences, Texas A&M University-Corpus Christi, Corpus Christi, Texas, USA
Submission: May 02, 2023;Published: May 12, 2023

Volume4 Issue5May, 2023
Abstract
Vibrio vulnificus is a gram-negative, halophilic bacterium normally found in temperate marine and estuarine waters. The organism may enter wounds in the skin and cause sepsis or necrotizing fasciitis; the latter condition has a 50-60% mortality rate and may result in death or disfigurement within 4-6 days. Persons exposed to coastal flood waters during hurricanes may be at risk for this organism. The virulence-correlated gene, vcgC, is specific for clinical isolates of V. vulnificus, but the function of this locus remains unknown. This study used Polymerase Chain Reaction (PCR) and novel primers for vcgC not previously described to specifically identify V. vulnificus isolates from the Texas Coastal Bend region that may result in serious infections. Of the 28 isolates, four isolates could not be revived multiple times; crude lysates of the remaining 24 Vibrio vulnificus cultures were analyzed by PCR, and 19 were found to have amplicons of 428bp for vcgC. This study confirms the presence of the vcgC gene in V. vulnificus isolates from the Texas Coastal Bend region of the Gulf of Mexico.
Keywords:PCR; Virulence factors; VcgC; Genome plasticity; Pathogenic marine bacteria
Abbreviations:ATCC : American Type Culture Collection, Manassas, VA,USA; N/A: Not Applicable; N/D: Not Done; WGS: Whole Genome Sequencing
Introduction
Vibrio vulnificus is a mesophilic, halophilic, asporogenous curved bacillus native to marine and estuarine waters. The organism may live in a planktonic state as single cells, or as a biofilm associated with the mucus on the skin of finfish and reside on the chitin exoskeletons of crustaceans, the calcareous shells of mollusks, or within the tissues of mollusks [1]. V. vulnificus has environmental importance as part of the marine ecosystem, but also major medical importance, as human hosts may contract gastroenteritis from raw or improperlycooked seafood [2], or may contract type III necrotizing fasciitis through wounds exposed to marine waters [2,3]. Recently, persons living in Florida who were exposed to flood waters resulting from Hurricane Ian in September 2022 and Hurricane Nicole in November 2022 reported much higher exposure to V. vulnificus. As of April 2023, the Florida department of health reported throughout the entire state of Florida 74 cases of vibriosis and 17 deaths in 2022 compared to 36 cases and seven deaths in 2020 and 34 cases and 10 deaths in 2021; the majority of cases were reported in Lee county, FL, with 28 cases and eight deaths [4]. This county was hardest hit by flood waters from Hurricane Ian. The overall figures for the state of Florida represent an increase in cases of 88% and a 70% increase in fatalities from vibriosis between 2021 and 2022. This pattern of increased numbers of vibriosis cases was also seen after Hurricane Katrina in Louisiana in 2005 [5]. The Coastal Bend region of south Texas is also prone to hurricanes, as seen with Hurricane Harvey in 2017. Flood waters from such storms may expose nearly a half-million inhabitants of the region to V. vulnificus, which is a concern for the large numbers of persons suffering from various pre-disposing conditions that increase susceptibility to vibrio wound infections such as diabetes, immunocompromising conditions including cancers, liver and kidney diseases, and hemochromatosis [6-8]. The incubation period for V. vulnificus wound infections ranges from three hours to 12 days, but symptoms appeared on average within 24 hours [7]. The time from wound inoculation to death ranged between three to nine days, but the average time was 4.5 days [6-8]. Clearly, rapid and accurate methods of identification of V. vulnificus in persons exposed to flood waters is critical to prevent the morbidity and mortality seen with V. vulnificus wound infections..
V. vulnificus may be characterized into three biotypes; most V. vulnificus infections in humans result from entry of biotype 1 [6]. Randomly amplified polymorphic DNA analysis revealed that V. vulnificus isolates were very heterogeneous [9], suggesting that other factors may confer virulence to this species. Subsequent investigations using traditional PCR described a locus unique to V. vulnificus biotype 1 isolates, titled the Virulence-Correlated Gene (vcg) [10], which has two allelic forms, vcgC and vcgE, both with functions remaining 3PCR Analysis of vcgC in Vibrio vulnificus unknown [11]. Strains possessing vcgC comprise 90% of clinical isolates, while strains with vcgE comprise 93% of environmental isolates [10]. Fermentation of mannitol was linked 100% to C-genotype strains, but only 40% of the time to E-type genotype [12]. In addition, the C-type showed increased resistance to human serum [13], indicating that this vcg genotype produces some unknown factors that provide increased ability to cause human infections. A later study indicated that C-genotype bacteria may have water and humans as a preferred host, while E-genotype may have oysters as a preferred host [14]. A recent report [15] compared predictive values, sensitivity and specificity of one phenotypic assay (D-mannitol fermentation) and three genotypic assay involving quantitative real-time PCR (16S rRNA of rrnA and rrnB genes, vcgC/E polymorphism and pilF polymorphism) of 171 environmental isolates and 81 clinical isolates. These bacteria were collected from South Carolina, Florida, Louisiana or Texas; or from Florida, Alabama or Texas, respectively. Their thorough study validated these four methods but did not list which gene loci of which individual isolates occurred at specific locations, especially the Coastal Bend region surrounding the city of Corpus Christi, which is the largest urban area situated on the Texas Gulf Coast, as Houston is located 50 miles or 80 kilometers from the Gulf. In addition, use of qPCR may not be plausible to implement following a hurricane due to destruction of laboratory infrastructure and inconsistent availability of electricity. Since little information was available on the vcgC locus, especially for Texas Coastal Bend isolates, devising primers using the vcgC locus might be more useful in distinguishing Vibrio vulnificus infections resulting from the floodwaters of hurricanes. This current investigation focused on the vcgC locus using traditional PCR, which can be performed by undergraduate students using basic thermocyclers and rudimentary gel electrophoresis equipment powered with basic generators. The purpose of this study was to design and use primers to identify vcgC within isolates taken from the coastal bend region. The primers described in this report are novel and have not been used in prior investigations. The study described herein suggests that vcgC alone may be a quicker method of analyzing pathogenic isolates of Vibrio vulnificus from this region, and the significance of this study is that these novel primers may be used under limited technical conditions (basic electricity, loss of more complicated real-time thermocyclers) to identify V. vulnificus isolates that may cause morbidity and mortality in persons exposed to hurricane flooding.
Method and Materials
Microorganisms
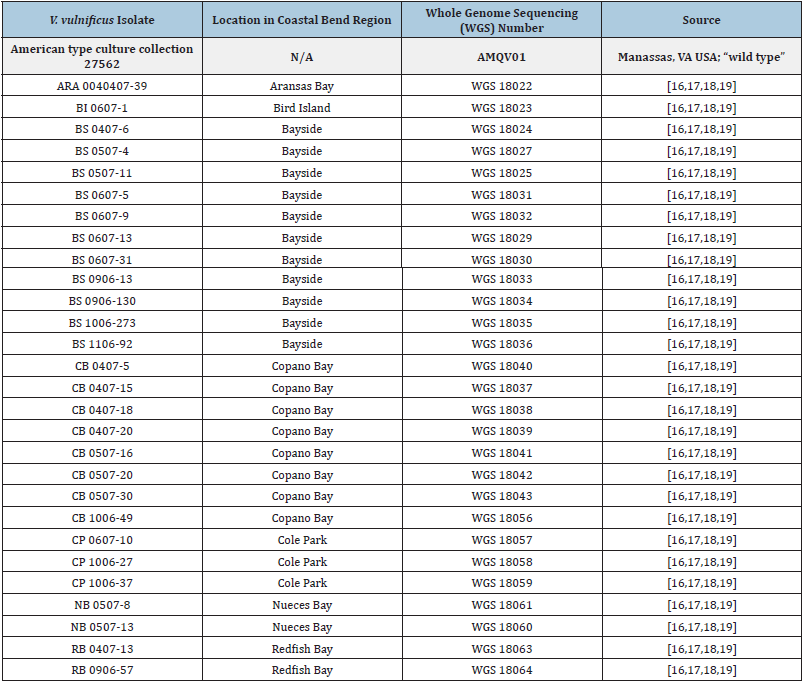
Strains were collected from the estuarine and marine waters of the Texas Coastal Bend and confirmed as V. vulnificus as originally described [16,17]. Additional characterization of the genomes as Vibrio vulnificus was performed as described previously [18,19]. Table 1 shows the 28 isolates; their genome sequences may be found in NCBI under bio project accession number PRJNA607302 [18,19].es will be chosen, and their socio-economic consequences [4,5].
Table 1: Vibrio vulnificus isolates used in this study.
Chemical media
Saturated overnight cultures of bacteria were grown in tryptic soy broth with 2% NaCl (TSB2N) or in Luria-Bertani Lennox broth with 2% NaCl (LBL2N). Cultures were preserved cryogenically at -80 °C in 100% vol/vol glycerol or 3% DMSO. Strains were revitalized by growing a 10μl loop of cryopreserved bacteria in Brain Heart Infusion Broth with 2% NaCl (BHIB2N), then sub cultured to 2mL of LBL2N and grown overnight at 35 °C to a concentration of ~109/ mL Bacteria were kept on solid agar slants of 7mL at ambient room temperatures using either Marine agar 2216 slants [20], Brain- Heart Infusion Agar slants with 2% NaCl, or Luria-Bertani Lennox agar slants with 2% NaCl and overlaid with sterile mineral oil.
Preparation of bacterial lysates
A crude lysate method was used to prepare bacterial samples [21]. Five hundred microliters of saturated overnight culture were washed in 1X TE buffer (100mM Tris-Cl, pH 8.0, 1mM EDTA, pH 8.0), centrifuged, heated at 100 °C, resuspended in 50μL of lysis solution(1mM EDTA, pH 8.0; 0.5% Triton-X100), re-heated at 100 °C for 10 minutes, then the supernatant kept and used for PCR. 4 PCR Analysis of vcgC in Vibrio vulnificus.
PCR primers
Primers sets were designed (Table 2) through publiclyavailable software packages. The Vibrio vulnificus gene sequence for vcgC described previously [10,22] and found as GenBank NC_005139 from V. vulnificus strain YJ016 [10,23], was used to design new primers. The current forward primer began annealing to accession number AY626575.1 from ATCC 27562 about 12 nucleotides downstream of the original sequence [23]. Primers were synthesized at 50nmol scale by Euro fins MWG/Operon (Huntsville, AL, USA), resuspended in 1X TE buffer (100mM Tris-Cl, pH 8.0, 1mM EDTA, pH 8.0), and aliquoted at -20 °C until use, then slowly thawed and diluted to 0.1-1.0μM in sterile deionized water just before use. The expected PCR product is an amplicon of 428bp.
Table 2: Vibrio vulnificus vcgC primers used in this study.

Generation and analysis of PCR amplicons
PCR reactions were performed in an Eppendorf Master cycler in 20μL reactions containing 2.0μL of 10X Roche Green PCR Mix with magnesium, 0.3μL of 10 mM dNTP solution (Roche), 1.0μL each of forward and reverse primers, 0.2μL of Roche Taq DNA polymerase, 14.2μL of sterile nano-pure water, and 2.0μL of bacterial lysate containing DNA. Reactions were done in duplicate at least three times at initial denaturation of 94 °C for 5 minutes, followed for 29 cycles at 94 °C for 40 seconds, 54 °C for 45 seconds, and 72 °C for 45 seconds with final extension at 72 °C for 10 minutes followed by 4 °C on hold. The PCR products were electrophoresed on 2% agarose gels containing 0.5μg/mL ethidium bromide using 1X Tris-acetate- EDTA and captured through gel detection system.
Result and Discussion
Initial attempts to detect Vibrio vulnificus isolates described in earlier reports were successful with vvhA and rpoS primers [24]. However, use of previous primers designed for vcgC [10,11,22] were unsuccessful, indicating either technical problems, or that all curated isolates contained the vcgE allele, or the primers were not specific enough for Texas Coastal Bend isolates. We hypothesized that redesigning primers for vcgC and for vcgE for detection of local isolates might be a logical step in differentiating whether the Vibrio vulnificus isolates were C-type or E-type. While attempts were made to develop multiplex PCR primers, the approach led to developing separate sets of vcgC and vcgE primers for separate PCR reactions. The original work collected 144 samples from local waters and isolated V. vulnificus from 142 samples [17] and use of a probe specific for vvhA confirmed 139 isolates as being V. vulnificus [16,17]. From these isolates, 42 strains that were cryo-preserved were saved after a loss of power from Hurricane Harvey in 2017, and these were subjected to whole-genome sequencing [18]. However, of these 42 isolates, only 28 isolates could be revitalized for this study. Growth of bacteria on marine agar 2216 slants, brain-heart infusion agar slants with 2% NaCl, and Luria-Bertani Lennox agar slants with 2% NaCl did not resurrect the other bacteria, plus four additional isolates did not grow, resulting in 24 isolates analyzed. The remaining 24 V. vulnificus isolates were cultured with the type strain ATCC 27562 and grown in duplicate in Luria-Bertani Lennox broth with 2% NaCl. Table 3 summarizes the results of the PCR analysis, and Figure 1 shows a representative 2% agarose gel with typical amplicons of 428bp, indicating the primers could amplify the vcgC gene. The results are summarized in Table 3.
Figure 1:The 2% agarose gel in 1X Tris-acetate -EDTA with 0.5μg/mL of ethidium bromide shows lane 1: Roche molecular weight marker VI; lane 2, BS 0607-5; lane 3, BI 0607-1; lane 4, ATCC 27562; lane 5, CB 0407-18; lane 6 is a positive control of 50ng of purified ATCC 27562 DNA, and lane 7 is a negative control with no DNA.
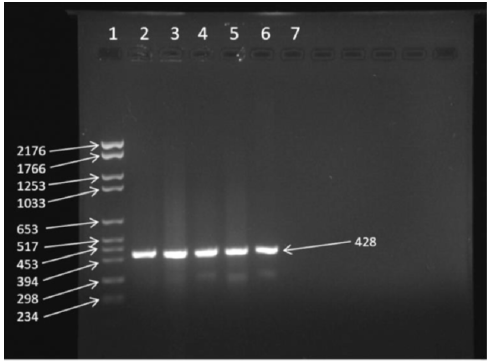
Table 3: Results of PCR for vcgC of Vibrio vulnificus isolates described here.
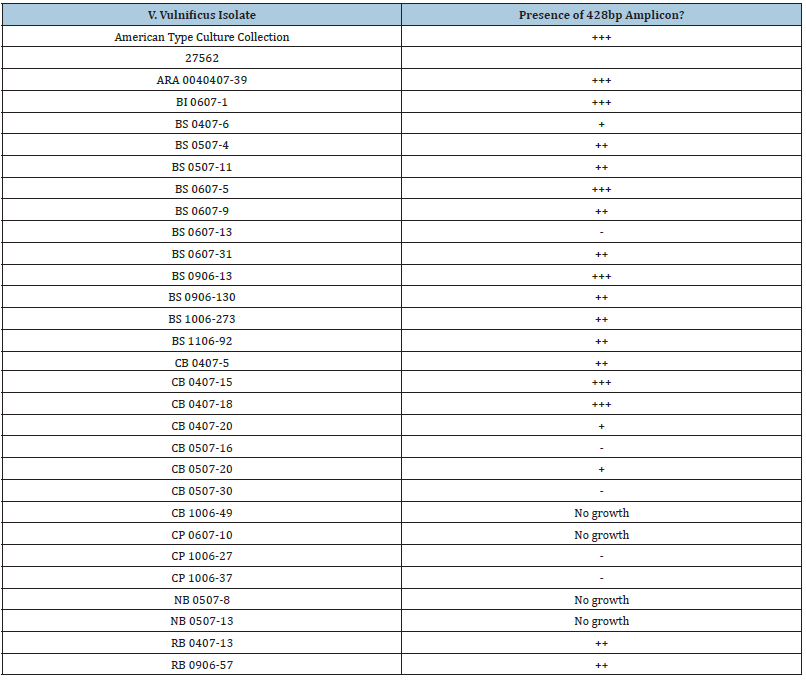
+=Yes, 428bp amplicon present for all reactions; ++ two of three reactions positive for amplicon +, one of three reactions positive for amplicon; -, no reactions revealed presence of 428bp amplicon.
Previous investigations designed vcgC primer pairs using sequences of vcgC from V. vulnificus isolates CMCP6 or YJ016 [10,22], which were found in Gen Bank under accession numbers NC_005139 from 2005 and NC_004459 from 2009, respectively. The primers used in this report were obtained from V. vulnificus isolate ATCC 27562 [23], accession number AY 626575.1; however, this latter sequence was characterized around 2012. A later investigation employed the identical primers described earlier [10] for Vibrio vulnificus isolates taken from a Mexico city seafood market [25]. The original description of quantitative real-time PCR vcgC primers were modified for endpoint PCR but were not successful due to either technical errors of 5 PCR Analysis of vcgC in Vibrio vulnificus neophyte undergraduates performing PCR for the first time, primer design in which a consensus sequence was not totally accurate in making the comparison between available sequences from the isolates that were described, or because the sequence had mutated. The first possibility was minimized by having groups of undergraduates in different cohorts repeating the PCR reactions in technical duplicate or triplicate samples, and also doing biological replicates, in which bacterial samples that were cryopreserved from different colonies of the original isolated samples [26]. The second possibility of an inaccurate consensus sequence cannot be excluded but decreasing stringency with decreasing salt concentration in the PCR buffer and decreasing melting temperature (Tm) should allow annealing of consensus sequences. Those approaches were not taken here, as the number of amplicons increased (data not shown). The third possibility of mutation is more likely; a report described that V. vulnificus genomes had a high level of plasticity [27]. This study found unique regions in both YJ016 and CMCP6 strains, called genomic islands, with the former strain possessing six unique regions not seen in 27 other strains that had been sequenced at that time [27]. This report also noted that prior investigations found a lack of PCR products with amplification, indicating that V. vulnificus isolates, regardless of their origin, “contain a large amount of strain-specific DNA” and all members of V. vulnificus characterized by whole genome sequencing thus far “possess genes with high variability” [27]. Additional studies in this lab confirm plasticity in the V. vulnificus genome, especially with designing primers for vcgE and multiplex vcgC/E primers [28,29].
Another recent report of comparisons between whole-genomes of both YJ016 and CMCP6 focused on the 237 pathogenicity associated genes from both strains and compared with two strains from Mexico, CICESE -316 and CICESE-325. The analysis showed that the Mexican isolates and strain YJ016 shared 63.2% of genes, while strain CMCP6 shared 63.9%. No mention was made of the variability with the vcgC locus between strains. A recent investigation found that use of three real-time PCR assays focusing on the 16S rRNA, vcg and pilF genes, coupled with a phenotypic method, fermentation of mannitol, accurately differentiated between 81 clinical and 171 environmental V. vulnificus isolates from different Gulf and Atlantic Ocean sites at a sensitivity averaging ~77% and a specificity of ~80% [15]. Their primers and probes for vcgC were described previously [22], and allowed for a characterization of 75.4% being vcgE, 15.8% being vcgC, with 8.8% unable to distinguish between the alleles. The 39 isolates from Texas waters between 2006 and 2007 may overlap with some of the isolates studied here, but information on specific strains was not available. This redesigned primer would be a good test on the measure of plasticity of the V. vulnificus genome, or if inability of primers to anneal is due to alterations in the genomic islands, insertion of phage genomes or transposable elements (R.R. Colwell, pers. comm, 2022). The work presented here provides additional information here on specific Texas Coastal Bend isolates.
Conclusion
The current study hypothesizes that specific redesign of vcgC primers using a more recent sequence of this locus would enable additional clarification of the 24 Texas Coastal Bend isolates and the vcgC locus. This new primer can detect vcgC in isolates from south Texas coastal waters, producing an amplicon of 428bp. The limitation is that additional work with vcgE primers is suggested in future studies to definitively categorize these south Texas isolates as C- or E- genotype. Coupled with confirmation of vcgC and previous whole genome sequencing information from the collection of 42 strains isolated from the Gulf of Mexico near Corpus Christi [18], and with previous works [10,15,18], analysis of these 24 isolates further elucidates what genes may be critical for V. vulnificus virulence. This study corroborates recent findings described by Dickerson et al. [15] on the use of the vcg locus to identify and characterize V. vulnificus isolates and also confirms an additional primer set for rapid PCR testing.
Acknowledgement
Author contributions
Conceptualization G.W.B; methodology G.W.B, S.D.G, and G.P.C; formal analysis G.B, L.I.B, G.P.C, and A.O.T; experiments G.W.B, L. I. B, J.S.C, G.P.C, S.D.G, T.B.O, D.S.P, A.O.T and G.D.R; cell curation G.W.B, G.P.C, and G.D.R; data curation, G.W.B, G.P.C; writing-original draft preparation G.W.B; writing-editing of draft G.W.B., G.P.C and G.D.R; project administration G.W.B, G.P.C; funding acquisition G.W.B, L.I.B, S.D.G, G.P.C, D.S. P, and A.O.T. 6 PCR Analysis of vcgC in Vibrio vulnificus.
Funding
This research was funded by NIH Extramural Associates Research Development Award PAR 05-053 to G.W.B; Texas A&M University-Corpus Christi University Research Enhancement Award to G.W.B.; Texas Research Development Funds Award to G.W.B. Undergraduate researcher L.I.B. was funded by the McNair Scholars Program to Texas A&M Univ-Corpus Christi. Undergraduate researchers S.D.G. and D. S. P. were funded by the Research Experience for Undergraduates to Texas A&M Univ.-Corpus Christi. G.P.C was funded by Bridging Master’s Students to the Doctorate, NIGMS 2R25 GM071936-034 subaward from the University of Texas Medical Branch; Undergraduate researcher A.O.T. was funded by the Texas A&M System Louis Stokes Alliance for Minority Participation. The lab of G.W.B. was presented a substantial gift by an anonymous donor. We thank Suzzette Chopin for additional support; Alyssa Garcia, Louise Lyle, Valerie May, Jessica Ramirez, Tyler Vance and Emilio Vasquez for technical assistance; Cherie McCollough for helpful comments to the manuscript. This manuscript is dedicated to Dr. Joanna B. Mott, former Chair of Life Sciences, who provided initial exposure to working with Vibrio vulnificus.
Conflict of Interest
G.W. B. has been an unpaid consultant for Jeevan Scientific, Dunwoody, GA USA. The company had no involvement in any aspects of this study. Work in the research laboratory of G.W.B. was also funded by an anonymous alumnus donor to Texas A&M University- Corpus Christi. G.W.B. is also a member of the National Board of Directors for the English-Speaking Union of the United States and may have inadvertent transient affiliations with donors or other persons indirectly affiliated with activities of said organization. G.W.B. will be receiving honorarium from the Louisiana State University Health Sciences Center at Shreveport and may have inadvertent transient affiliations with editors or reviewers from that institution.
References
- Thompson FL, Iisa T, Swings J (2004) Biodiversity of vibrios. Microbiol Mol Biol Rev 68(3): 403-431.
- Hlady WG, Klontz KC (1996) The epidemiology of Vibrio infections in Florida, 1981-1993. J Infect Dis 173: 1176-1183.
- Tsai YH, Hsu RWW, Huang KC, Huang TJ (2011) Comparison of necrotizing fasciitis and sepsis caused by Vibrio vulnificus and Staphylococcus aureus. J Bone Joint Surg Am 93(3): 274-284.
- http://www.floridahealth.gov/
- Rhoads J (2006) Post-hurricane katrina challenge: Vibrio vulnificus. J American Academy of Nurse Practitioners 18(7): 318-324.
- Oliver JD (2005) Wound infections caused by Vibrio vulnificus and other marine bacteria. Epidemiol Infect 133(3): 383-391.
- Horseman M, Surani S (2011) A comprehensive review of Vibrio vulnificus: An important cause of severe sepsis and soft-tissue infection. Intl J Inf Dis 15(3): e157-e166.
- Klontz, KC, Lieb S, Schreiber M, Janowski HT, Baldy LM, et al. (1988) Syndromes of Vibrio vulnificus infections. Clinical and epidemiologic features in Florida cases, 1981-1987. Annu Int Med 109(4): 318-323.
- Warner JM, Oliver JD (1999) Randomly amplified polymorphic DNA analysis of clinical and environmental isolates of Vibrio vulnificus and other Vibrio species. Appl Env Microbiol 65(3): 1141-1144.
- Rosche TM, Yano Y, Oliver JD (2005) A rapid and simple PCR analysis indicates there are two subgroups of Vibrio vulnificus which correlate with clinical or environmental isolation. Microbiol. Immunol 49(4): 318-389.
- Rosche TM, Binder EA, Oliver JD (2010) Vibrio vulnificus genome suggests two distinct ecotypes. Environmental Microbiology Reports 2(1): 128-132.
- Froelich BE, Oliver JD (2011) Orientation of mannitol-related genes can further differentiate strains of Vibrio vulnificus possessing the vcgC allele. Adv Studies in Biol 3(4): 151-160.
- Bogard RW, Oliver JD (2007) Role of iron in human serum resistance of the clinical and environmental Vibrio vulnificus genotypes. Appl Environ Microbiol 73(23): 7501-7505.
- Warner E, Oliver JD (2008) Population structures of two genotypes of vibrio vulnificus in oysters (Crossostrea virginica) and seawater. Appl Environ Microbiol 74(1): 80-85.
- Dickerson J, Gooch MJ, Jacobs JM, Mott JB (2021) Characteristics of vibrio vulnificus isolates from clinical and environmental sources. Molecular and Cellular Probes 56: 101695.
- Ramirez GD (2008) MS thesis. Texas A&M University-Corpus Christi, Corpus Christi, Texas, USA.
- Ramirez GD, Buck GW, Smith AK, Gordon KV, Mott JB (2009) Incidence of vibrio vulnificus in estuarine waters of the south Texas Coastal Bend region. J Appl Microbiol 107(6): 2047-2053.
- Mullis MM, Huang IS, Planas CGM, Pray R, Buck GW, et al. (2019) Draft genome sequences of 42 environmental Vibrio vulnificus strains isolated from the Northern Gulf of Mexico. Microbiol Resour Announc 8(28): e00200-e00219.
- Planas CGM, Mullis MM, Huang IS, Buck GW, Turner L, et al. (2018) Genome-scale phylogenetic analysis of environmental Vibrio vulnificus from the Texas Coastal Bend region of the northern Gulf of Mexico. Texas Branch of the American Society of Microbiology meeting, Corpus Christi, Texas, USA, pp. 8-10.
- Gomez GB, Roque A (2006) Isolation, enumeration, and preservation of the Vibrionaceae. In: Thompson FJ, Austin B, Swings J (Eds.), Biology of Vibrios, ASM Press, Washington, USA, pp. 15-28.
- Parvathi A, Kumar HS, Karunasagar I, Karunasager I (2005) Study of the occurrence of Vibrio vulnificus in oysters in India by polymerase chain reaction (PCR) and heterogeneity among V. vulnificus by randomly amplified polymorphic DNA PCR and gyrB sequence analysis. Envir Micro 7(7): 995-1002.
- Baker AC, Gore A, Oliver JD, Rangdale R, Mc Arthur JV, et al. (2010) Rapid in situ detection of virulent Vibrio vulnificus strains in raw oyster matrices using real-time PCR, Environ Microbiol Rep 2 (1): 76-80.
- Li Z, Chen H, Chen X, Zhou T, Zhao L, et al. (2012) Genome sequence of the human pathogenic bacterium Vibrio vulnificus type strain ATCC 27562. J Bacteriol 194(24): 6954-6955.
- Planas CGM (2014) MS thesis. Texas A&M University-Corpus Christi, Corpus Christi, Texas USA.
- Guerrero A, Gil RBG, Wong CI, Lizarraga PML (2015) Genetic characterization of Vibrio vulnificus strains isolated from oyster samples in Mexico. Int J Environ Health Res 25(6): 614-627.
- Bell G (2016) Replicates and repeats. BMC Biology 14: 28.
- Quirke AM, Reen FJ, Claesson MJ, Boyd EF (2006) Genomic-island identification in Vibrio vulnificus reveals significant genome plasticity in this human pathogen. Bioinformatics 22(8): 905-910.
- Sama V (2021) MS thesis. Texas A&M University-Corpus Christi, Corpus Christi, Texas USA.
- Guerrero A, Licea NAF, Gonzalez SR, Lizarraga PML (2019) Whole-genome comparison between reference sequences and oyster Vibro vulnificus C-genotype strains. PLOS One 14(7): e0220385.
© 2023 Gregory W Buck. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






