- Submissions

Full Text
Journal of Biotechnology & Bioresearch
Wine Yeast Communities Observed in Spontaneous Fermentations of Five Greek Grape Varieties Selected from Five Greek PGI Regions
Aikaterini Karampatea*
Department of Agricultural Biotechnology and Oenology, International Hellenic University, Greece
*Corresponding author:Aikaterini Karampatea, Department of Agricultural Biotechnology and Oenology, International Hellenic University, Greece
Submission: January 05, 2023;Published: January 27, 2023

Volume4 Issue3January , 2023
Abstract
Microbiological cultures were isolated from Greek grapes and musts subjected to spontaneous alcoholic fermentations on a micro-vinification scale. After the isolation and purification of the cultures, was performed phenotyping of 250 isolates by combining culture on Chromogram chromogenic medium and using the API ID32C identification system and study of phenotypic characteristics. At the molecular level, PCR-Delta and PCR-ITS-RFLP techniques were applied due to their ability to distinguish S. Cerevisiae yeasts from Non Saccharomyces as well as differentiation of Non Saccharomyces species using two restriction enzymes (HinfI and HaeIII). Among the isolated yeasts apart S. Cerevisiae, for the rest of the yeasts there is no statistically significant correlation with any of the study regions. There seems to be some association between the Assyrtiko variety and Kluyveromyces thermotolerance species, regardless of place and cultivation method.
Keywords: Yeasts; Kluyveromyces; Hydrogen sulfide; Grapes; S. Cerevisiae
Introduction
Studies on more or less geographically limited areas and with various molecular methods attempting to find explanations for the discrepancies found in studies on the issue of terroir strains [1]. Indeed, even if a microbial signature could be found, the effect of a species (eg Non- Saccharomyces) present at concentrations a hundred or thousand times lower than that of the main contributor, i.e. S. Cerevisiae, remains to be determined [1]. Such studies should also allow tracking the distribution of floras across geography and over time, and possibly attempt to understand the evolution that creates their great diversity.
Non-Saccharomyces yeast species on the surface of grapes vary according to the stage of grape ripening. From fruit set to ripening, a succession of Non-Saccharomyces yeasts is observed. Regarding S. Cerevisiae yeasts, according to some researchers, they are undetectable in grapes [2,3] while other studies have shown that the species S. Cerevisiae can be found in grapes and in some cases in sufficient proportion to cause the initiation of alcoholic fermentation [4].
Different grape microbial communities can be influenced by various factors, such as for example the use of phytosanitary products at the stage of ripening of the grapes [5] or the organic or biodynamic management of farms based on copper-based fungicides [6-7]. According to Patrignani et al. [8] yeast identification analyzes performed on musts from biodynamically grown grapes, under spontaneous fermentation after 5, 8, 10 and 11 days of fermentation showed a higher resistance of Non-Saccharomyces species in relation to the fermentation must from organic grapes. After 10 days of fermentation S. Cerevisiae represented the main species found in the biodynamic wort, while in the organic wort, S. Pombe was at the highest level [8].
An important preliminary step in the selection of yeast strains for their use as starters in alcoholic fermentations is that they meet a number of oenological characteristics, such as low production of hydrogen sulfide, ability to ferment at high temperatures, tolerance to stressful conditions (concentration of alcohol, sugars, sulfur dioxide), enzymatic actions, fermentation capacity. In addition, the chemical profile of the produced wines (with a number of parameters including total, active and volatile acidity) and the aromatic profile are also of decisive importance, as the various strains affect the chemical composition and organoleptic characteristics of a wine by producing different amounts of secondary products. We must not forget that the wines.
Whether varietal-specific microbial communities actually regulate the organoleptic properties of wine should be tested experimentally. How climate change, which is more pressing than ever, affects microbial community responses should be studied. Essentially through the above research it is as if an opportunity is presented for the development of adapted techniques that will contribute to the improvement of the quality of grape varieties and wine.
Material and Methods
Sources of yeast isolates: For our study, five Greek grape varieties (Malagousia, Assyrtiko, Vidiano, Moscofilero, Agiorgitiko), with particular oenological importance, were selected from five different wine-growing protected geographical indication regions of Greece (Drama, Kavala, Thessaloniki, Halkidiki, Atalanti) with conventional or organic farming 25kg of healthy grape bunches was collected using sterile scissors, stored in sterile bags in the freezer (-18 ˚C). For the production of must, fresh or thawed grapes were crushed by hand 20g/hL (NH4)2SO4 was added to the must, left for spontaneous fermentation at 25 ˚C. All spontaneous experimental fermentations were carried out in temperature-controlled 30L stainless steel tanks, faithful replicas of professional winemaking tanks (Figure 1).
Figure 1:Maps showing grape sampling areas (google.com/maps accessed 04/02/2022)
Isolation, purification and preservation of cultures: Native
yeast flora was isolated a) from grapes, and b) in 3 phases of
spontaneous fermentations:
i) beginning (12-14oBe)
ii) middle (6oBe)
iii) end (<1oBe)
For isolation, 1mL of the grape wash or must, after serial dilutions, was added to nutrient medium (YPD, Sigma-Aldrich, USA) supplemented with streptomycin sulfate (250mg/L) (Fisher Scientific Belgium) followed by incubation for 5 days at 25 ˚C [9-11]. Plates containing 30-300 colonies were examined macroscopically followed by re-plating of isolated colonies on YPD agar.
Microbiological identification
Phenotypic identification: The isolated yeast colonies were first identified phenotypically [12]. Fermentation of the sugars galactose, sucrose, maltose, raffinose, meliviose and starch was also carried out in test tubes containing 10mL of synthesis substrate: peptone 1%, yeast extract 0,5%, Phenol-red 0.00024% [13] with the simultaneous addition of a 2% agar layer 2cm thick on the surface, in order to achieve anaerobic conditions [14]. The aqueous sugar solution was sterilized by filtration through 0.22μm filters (Millipore, France) and an appropriate amount was added to the test tubes to achieve a final concentration of 2%v/v. Then the test tubes were inoculated with 0.1mL of an aqueous suspension of cells of the strain to be examined, incubated at 25 °C for a period of 14 days and visually examined for any color change of the substrate and/or production of carbon dioxide indicating fermentation of the sugar to be examined.
Chromagar:Chromagar (agar 15g/L, peptone 10.2g/L, chloramphenicol 0.5g/L, chromogenic mixture 22g/L, pH 6.1±0.2) is a chromogenic differential culture medium that facilitates the isolation and possible identification of certain of clinically important yeast species, especially when in the YPD medium the differentiation is not visible [15].
ID 32C detection system: The ID 32 °C detection system (Bio- Merieux, France) was used for the carbon assimilation assays. ID 32 °C is a standardized system for yeast identification, which uses 32 microscopic assimilation tests (dehydrated carbohydrate substrate) and a database.
Molecular characterization of yeast cultures:Strains were sub cultured in 5mL of YPD broth followed by overnight incubation under agitation at 30 °C. Then, genomic DNA was isolated using a commercial kit (Genomic DNA from tissue, Macherey-Nagel, USA), according to the relevant protocol (Support protocol for yeast 01/2017, Rev.17). Finally was obtained 50μL of mixture containing 5-30ng/μL genomic DNA. DNA amplification was performed in a final volume of 25μL containing 0.2mM dNTP (Invitek, Germany), 1.25μL of each primer (100 pmol/μL) (Kapa Biosystems, USA), PCR reaction buffer (1X) and 0.1μmol/min Taq DNA polymerase (KAPA2G Robus) (Kapa Biosystems, USA). PCR conditions were as follows: initial cycle of denaturation at 95 ˚C for 5min followed by 35 cycles of amplification, denaturation at 95 ˚C for 15sec, recovery at 55 ˚C for 15 sec and extension at 72 ˚C for 30sec and final extension at 72 ˚C for 10min. The amplification reaction was carried out in PCR Mini Opticom (Bio-Rad, France). PCR fragments were separated and detected by agarose gel electrophoresis (1.5 %) in TBE buffer (1X) at 120 V for 2.5h (Sub-Cell GT Agarose Gel Electrophoresis Systems, Bio-Rad, France). The gel was stained with gel red (10mg/L) and visualized under UV light (Gel Doc EZ Imager, Bio-Rad France) with Image Lab software (Bio-Rad, France). For RFLP analysis, PCR products were purified by ethanol precipitation followed by incubation with restriction endonucleases HaeIII, HinfI, (Takara, Japan) following the manufacturer’s instructions. Restriction fragments were separated on an agarose gel (1.5%) under the same conditions as the amplified products. Representative samples were clustered by PCR-RFLP of the ITS regions.
PCR-RFLP (ITS1-ARNr 5.8S-ITS2):Internal transcribed spacers (ITS) (ITS1 and ITS2) and 5.8S rDNA gene regions were amplified using specific primers ITS1 (5_-TCCGTAGGTGAA CCTGCGG-3_) and ITS4 (5_-TCCTCCGCTTATTG ATATGC-3_) [15].
PCR-Delta (Delta 12/Delta 21):The typing of Saccharomyces cerevisiae strains was carried out by PCR, according to the method developed by Legras and Karst [16] with primers: δ 12(5’- TCAACAATGGAATCCAAC-3’) and δ 21(5’-CATCATTAACACCCGTATATGA-3’). After amplification, Delta PCR profiles were obtained by electrophoresis for 40min at 100- 120V.
Oenological criteria:The ability of strains to grow at various temperatures was determined by visual observation [17,18]. Fermentative capacity was determined in the absence/presence of sulfuric anhydride. Fermentation rate was expressed as weight loss due to CO2 production (gCO2 per 100mL) after 2 and 7 days (Monaco et al., 2014). The flocculation properties of selected strains were tested according to Bony et al. (1997) and Suranka et al. [19]. β-glucosidase production was tested according to Strauss et al. [20] Colonies with glucosidase activity were identified due to the clear zone around the colonies [20,21]. Ethanol tolerance was studied according to Parish [22]. Somnolence of selected strains was tested in 5 mL of YPD medium with 40% (w/v) and 50% (w/v) glucose [19]. Malic and acetic acid production by selected strains, according to Pretorius [23]. Hydrogen sulfide production was studied on Bismuth Sulfide agar (BIGGY agar) [24,25]. The API-ZYM system (Bio-Merieux, France) was used for the semiquantitative examination of the following 19 enzyme reactions: alkaline phosphatase, esterase, esterase/lipase, lipase, leucine, valine, cystine, trypsin, chymotrypsin, phosphatase, phosphorylase, α-galactosidase, β-galactosidase, β-glucuronidase, α-glucosidase, β-glucosidase, glucosaminidase, α-mannosidase, α-fructosidase.
Result and Discussion
In our study, we found that grapes of Malagousia and Assyrtiko varieties are a rich source of yeasts, as four different types of yeasts (Saccharomyces and Candida, Pichia, Kloeckera) were isolated. Overall, significant differences are maintained between the different grape varieties in terms of the qualitative and quantitative distribution of yeast species, since for the Assyrtiko variety cultivated in 3 different viticultural zones, Kluyveromyces thermotolerance yeast was isolated at the beginning and in the middle of fermentation in grapes from vineyards of conventional and organic farming.
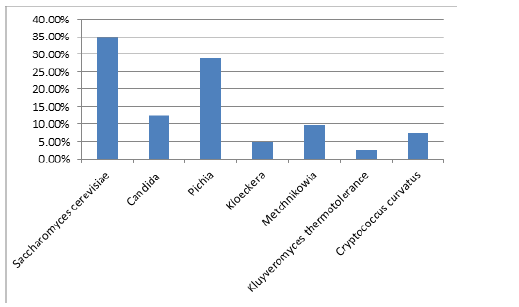
Among the 250 isolates isolated from different grape varieties, 87 yeast isolates belong to the species Saccharomyces cerevisiae (34.80%), the remaining isolates represent Non-Saccharomyces species and have been identified as belonging to different genera including Pichia (28.80%). Candida (12.40%), Metchnikowia (9.60%), Cryptococcus curvatus (7.20%), Kloeckera apiculata (4.80%) and Kluyveromyces thermotolerance (2.40%).
Many microorganisms are inhibited by M. Pulcherrima, including Candida tropicalis, Candida albicans, Brettanomyces/ Dekkera, Hanseniaspora, Pichia, and Botrytis cinerea. The yeast S. Cerevisiae appears to be unaffected by this antimicrobial activity. Additionally, some strains of M. pulcherrima have the ability to arrest the growth of susceptible organisms. M. pulcherrima is also described as a bio fungicidal agent that can reduce the population of the fungus B. cinerea in fruits, during the feeding process [26].
Additionally, generally in spontaneous fermentations there is a sequential succession of yeasts. First, the species Kloeckera, followed by Rhodotorula, Pichia, Candida, Metschnikowia and Cryptococcus which are found at low levels in fresh must [2,22,27-30]. Of these, the species H. Uvarum is usually present in higher population, followed by various Candida spp.[23] (Figure 2). In Table 1 are shown the means and standard deviations of the yeasts in each region. S. Cerevisiae is found more in the area of Kavala (Mean=.346 SD=.154), while in the area of Atalanti is found less (Mean=.020 SD=.045). At the same time, Cryptococcus yeast is found more in the area of Kavala (Mean=.154 SD=.226), while in the areas of Thessaloniki (Mean=.036 SD=.081) and Atalanti (Mean=.036 SD=.081) is found less. In addition, S. Kluyveri is found more in the area of Drama (Mean=.110 SD=.246), while in the areas of Thessaloniki and Atalanti is found less (Mean=.00 SD=.00). As for C. globosa, is found more in the area of Halkidiki (Mean=.166 SD=.235) and less in Atalanti and Thessaloniki (Mean=.00 SD=.00). C. Lusitanea, it is found more in the area of Kavala (Mean=.246 SD=.369), while in the area of Thessaloniki is found less (Mean=.00). At the same time, yeast C. Colliculosa is found only in the region of Halkidiki (Mean=.100 SD=.224) and not at all in the other regions. Regarding Pichia spp., is found more in the area of Kavala (Mean=.166 SD=.163) and less in the area of Thessaloniki (Mean=.016 SD=.036). Regarding the yeast Kloeckera apiculata, is found more in the regions of Halkidiki and Drama (Mean=.100 SD=.148) and less in the region of Thessaloniki (Mean=.034 SD=.076). At the same time, Metchnikowia is found more in the area of Kavala (Mean=.206 SD=.205), while in the area of Thessaloniki and Atalanti is not found at all (Mean=.00). Finally, the yeast Kluyveromyces thermotolerance is found more in the area of Kavala (Mean=.100 SD=.224), while in the areas of Thessaloniki and Atalanti is not found at all (Mean=.00).
Figure 2:Frequencies of yeast genera isolated from the 5 grape varieties in 5 viticultural zones.
From Table 1 and Figure 3 we observe that the yeast S. Cerevisiae is found more in Kavala. At the same time, according to Table 2 significant correlations are found between the areas of Kavala-Thessaloniki (Sig.=.014<.05 Mean Difference=.316), as well as between Kavala and Atalanti Sig.=.011 Mean Difference=.326).
Table 1: Means and standard deviations of yeast species for each region.
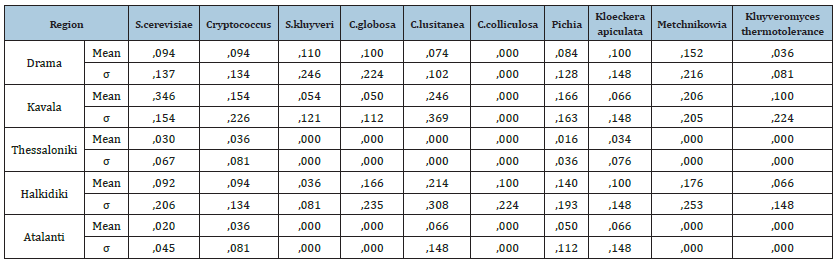
Figure 3:Depiction of the yeast S. cerevisiae in all study areas.
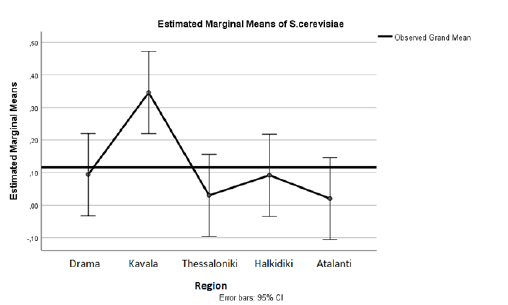
Table 2: Associations of the yeast S .cerevisiae with the study areas.
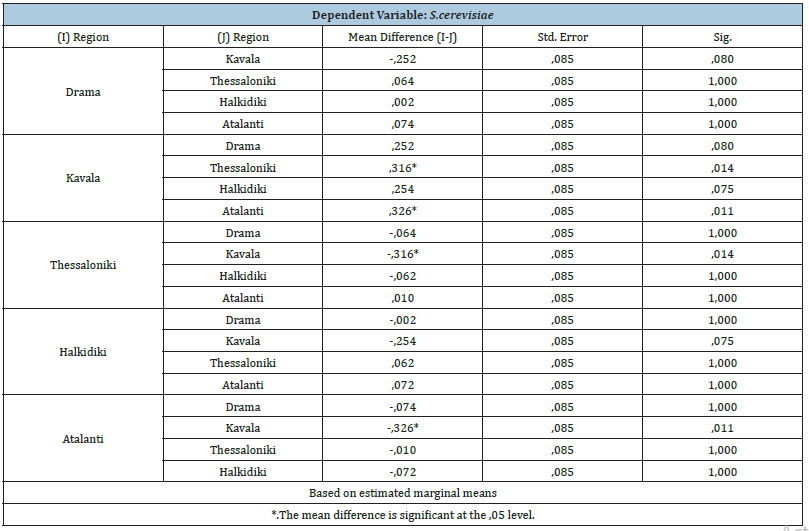
For the rest of the yeasts, there is no statistically significant
correlation with any of the study areas, as the results of the
correlation of each type of yeast with the study areas give us the
following results:
Cryptococcus (Sig.=.663>.05).
S. Kluyveri (Sig.=.642>.05).
C. Globosa (Sig.=.399>.05).
C. Lusitanea (Sig.= .408>.05).
C. Colliculosa (Sig.=.431>.05).
Pichia (Sig.=.419>.05).
Kloeckera apiculata (Sig.=.932>.05).
Metchnikowia (Sig.=.209>.05).
Kluyveromyces thermotolerance (Sig.= .668>.05).
For Malagousia and Assyrtiko varieties, where the largest volume of sampling was carried out, we had a larger number of vine plots cultivated either conventionally or organically, and in most cases a three-year study (Table 3).
Table 3:Percentage distribution of S. cerevisiae and Non-Saccharomyces yeasts, by variety in general and by variety and cultivation method.
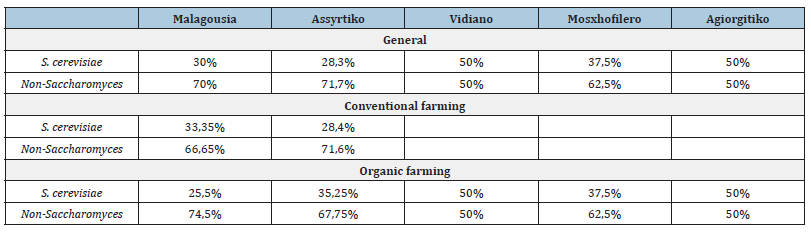
Malagousia
For the Malagousia variety, in 4 out of 6 vineyard plots of conventional cultivation, regardless of region, S. Cerevisiae was isolated from the grapes, as well as Candida colliculosa and Pichia, regardless of region and cultivation. Kloeckera apiculata was isolated in only one vineyard plot of conventional cultivation in the region of Atalanti. Regions of Drama and Halkidiki have the greatest biodiversity. The 2 study vineyard plots in Afitos Halkidiki were organically farmed. Results that are in agreement with studies that conclude that organic farming increases both the abundance and richness of biodiversity species (https://read. organicseurope.bio/ publication/organic-farming-and-biodiversity/pdf/).
Assyrtiko
In 3 of the 4 observation vineyard plots, Pichia yeasts were isolated from the grapes (conventional and organic vines), in 2 of the 4 Candida globosa (conventional and organic vines) and only in one of them Kloeckera apiculata. However, the most important finding is that the yeast Kluyveromyces thermotolerans was identified in all 3 study viticultural zones. Something observed only for the Assyrtiko variety.
In relation to Kluyveromyces thermotolerans yeasts have been detected by many researchers [31-33] in grapes and musts in many wine-growing regions around the world, including Greece. Kluyveromyces thermotolerans have been observed to be able to show further tolerance to ethanol and survive to the end of the fermentation course [31,33]. Data that are in agreement with what we observed in the Assyrtiko variety.
It is also interesting that using grapes harvested in aseptic conditions and immediately fermented under sterile conditions, in 68% of the samples fermentation started, of which 42% were complete and 28% were dominated by the yeast S. Cerevisiae [34]. In another study based on aseptic handling of grapes, the main species observed during fermentation are T. Delbrueckii and S. Cerevisiae (Clemente et al., 2004). Very important details since in our study the same protocol regarding aseptic conditions was applied to the sampling/harvesting of the grapes and the vinification’s (Table 4).
Table 4:Percentage distribution of S. cerevisiae and Non-Saccharomyces yeasts, by sampling area.
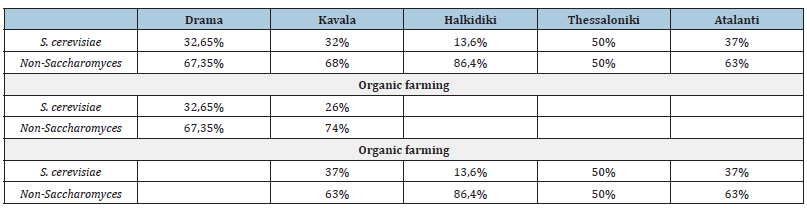
Despite their important role in alcoholic fermentation, Saccharomyces cerevisiae was, in most cases, not detected on grape surfaces, being isolated only from must. These data are in agreement with previous work, which reports that S. Cerevisiae is not a dominant species on grape surfaces and at the beginning of fermentation, but that it dominates from the middle to the end of fermentation [35]. Its incidence may also vary as a function of geographic region and grape variety [36].
Changes of yeast species diversity and their population were observed during all spontaneous alcoholic fermentations. Alcoholic fermentation of grape must mainly involves the development and activity of various species and strains of yeast, contributing to the complex chemical composition and organoleptic properties of wine. Understanding changes in the diversity and population of yeast flora is important for winemakers to control alcoholic fermentation and thus wine quality [37].
Table 5 presents the characteristics of the environmental conditions of the sampling areas indicatively for the same year. We notice that the regions of drama and Kavala have almost the same average high and low temperatures, common characteristics of rainfall (frequency, amount) as well as winds. Their only difference is the distance from the sea and the fact that the sampling plot in Koinonia of Drama is surrounded by three mountain ranges (Falakro, Menoikio, Pangaio). These data combined with the data in Table 4 may explain the similar rate of occurrence of S. Cerevisiae and Non-Saccharomyces yeasts in these 2 sampling areas.
Table 5:Characteristics of the environmental conditions of the viticultural sampling areas (www.meteoblue.com, www. apostaseis.gr accessed 04/02/2022).
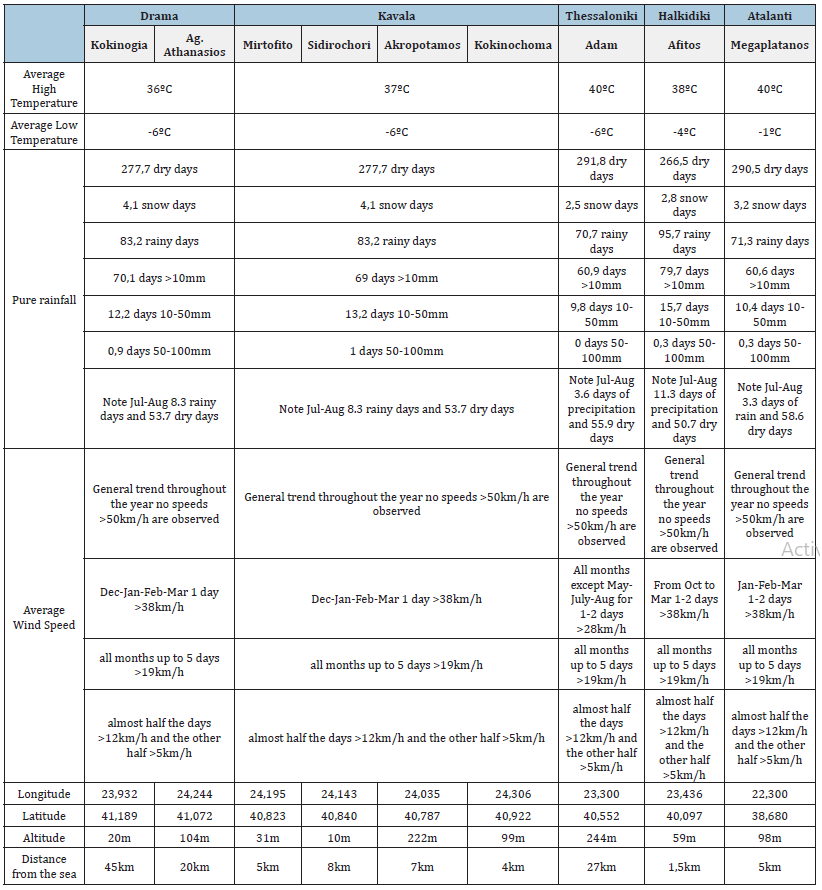
There are studies that found no differences in the composition and dynamics of yeast species during fermentation [38] or in the main yeast species of grapes from different Vitis vinifera cultivars [39]. In contrast, Raspor et al. [3] described some variability among yeast species (mainly Non-Saccharomyces) from different grape varieties and locations, but found no effect of grape variety on yeast engraftment [40] González-Alonso I et al. (2021) report for the first time the isolation and identification of four Non- Saccharomyces yeasts (Metschnikowia pulcherrima, Kluyveromyces thermotolerance, Hanseniaspora uvarum and Torulaspora delbrueckii) from grapes and must of the Negro Saurí variety. A result similar to previous reports on the microflora associated with Prieto Picudo, Carinyena and Garnacha varieties from the Priorate wine region of Tarragona in Spain. Also according to Banilas G. et al. [32] it is not yet clear if the concept of microbial terroir can also apply to Non-Saccharomyces wine yeast species. Their research resulted in a defined biogeography for Kluyveromyces thermotolerance in Greece. Despite the intra-regional genetic variation of the isolates, the genetic structure between the populations of the Nemea and Pea viticultural zones coincides with distinct oenological phenotypes, a necessary condition for the microbial contribution of terroir to wines [32].
Karyotype analyzes were also performed on Saccharomyces cerevisiae strains isolated at different stages of fermentation. A small number of Saccharomyces cerevisiae strains were found to be able to dominate alcoholic fermentation, regardless of grape variety and region. Similarly, stability was observed among dominant yeast karyotypes. As well as that at the end of the fermentations, in almost all cases, Saccharomyces cerevisiae was detected in the majority.
However, understanding the underlying causes of phenotypic variation and linking it to molecular determinants, especially for often complex quantitative traits, is not always easy. Microbial diversity has been suggested as an important indicator of soil quality and ecosystem stability [41,42]. Reports of the effects of fertilization, crop rotation, and pesticide application on soil microbial diversity as measured by parameters such as richness, relative abundance, and distribution vary among different studies [43,44], and the sensitivities of these parameters in response to environmental influences are largely unknown and most studies have focused on bacterial diversity. Changes in microbial community structures may not necessarily lead to altered diversity because changes in some taxa may be offset by changes in others. It has been suggested that, for example, species richness may show less variability in response to environmental factors than species composition [45].
The five viticultural zones selected for the study are characterized by different topography, variability of soil types, climates, microclimates (five different zones of protected indication) but also present a great variety in cultivated varieties. In relation to the applied viticultural practices, these were all the same, apart from the conscious choice to cultivate some plots of land under the rules of integrated management and others according to the rules of organic farming.
Geography is one of the first factors that can explain microbial population structures, as seen for plants [46] and we tried to see if this factor had an effect in our study as well, as the distances of vineyard plots could be adjacent or 40km apart for the same zone or even 540km between different viticultural areas. The differentiation does not seem to be related to the distance between the sites, but perhaps to natural barriers (mountain ranges) as in the case of the Malagousia plot in the area of drama in Koinonia.
With the exception of the yeast S. Cerevisiae, for the rest of the yeasts there is no statistically significant correlation with any of the study areas. The yeast S. Cerevisiae was found more in Kavala than in the rest of the regions. It was also observed that within each viticultural zone, there are differences in the S. Cerevisiae yeast populations depending on the subzone. Along with these natural environmental parameters, man with his action can influence the diversity of yeasts. The choice of vineyard management method, which mainly depends on the plant protection protocol applied, can potentially affect the microbial community [47,48] and in particular the diversity of the yeast S. Cerevisiae.
However, the results obtained during three years of study show a higher diversity of S. Cerevisiae strains for the plots of conventional agriculture than those of organic agriculture. Perhaps the types of fungicides used, their modes of application and/or actions (contact, surface or systemic, by translocation) firstly affect the structure of yeast communities of grape berry surfaces and secondly their properties. In our research results there is evidence that microbial biogeography is related to the grape at the varietal level and is also non-randomly related to local, varietal and climatic factors in viticultural zones.
Also, a rotation of dominant strains was observed from one year to the next, suggesting that these strains are not necessarily better adapted than others to establish and carry out fermentations over several years, but that their appearance or dominance depends from the conditions that prevail in each season. In studies it has been observed that indigenous S. Cerevisiae strains occurring in higher percentages in spontaneous alcoholic fermentations are more competitive, possibly due to their higher adaptability to the natural environment as well as to the progressive changes of environmental conditions during fermentation, especially the content in ethanol and temperature [49,50]. Also, it has been observed that for the same winery some dominant strains of S. Cerevisiae persisted in different fermentations from one year to another and appear to be representative of a single winery rather than an oenological geographic region [51-55].
Whether varietal-specific microbial communities actually regulate the organoleptic properties of wine should be tested experimentally. How climate change, which is more pressing than ever, affects microbial community responses should be studied. Essentially through the above research it is as if an opportunity is presented for the development of adapted techniques that will contribute to the improvement of the quality of grape varieties and wine [55,56].
Conclusion
An important preliminary step in the selection of yeast strains for their use as starters in alcoholic fermentations is that they meet a number of oenological characteristics, such as low production of hydrogen sulfide, ability to ferment at high temperatures, tolerance to stressful conditions (concentration of alcohol, sugars, sulfur dioxide), enzymatic actions, fermentation capacity. In addition, the chemical profile of the produced wines (with a number of parameters including total, active and volatile acidity) and the aromatic profile are also of decisive importance, as the various strains affect the chemical composition and organoleptic characteristics of a wine by producing different amounts of secondary products. We must not forget that the wines produced are intended for human consumption and must have desirable characteristics for consumers.
As a consequence of all the above, we consider that it would also be appropriate to carry out tests on various Greek grape varieties, in order to study the possible contribution of the “Greek” strains to the typicality of the wines produced in Greece. Also, it would be interesting to verify their importance by applying different vinification protocols, but also their adaptation to the vinification of different varieties by applying the same and/or different vinification protocols, as well as the results of their comparative application with commercial yeast strains that are already on the market.
We also encourage further studies on more or less geographically limited areas and with various molecular methods that we speculate will help to find explanations for the discrepancies found in studies on the issue of terroir strains [1]. Indeed, even if a microbial signature could be found, the effect of a species (e.g., Non- Saccharomyces) present at concentrations a hundred or thousand times lower than that of the main contributor, i.e. S. Cerevisiae, remains to be determined [1]. Such studies should also allow tracking the distribution of floras across geography and over time, and possibly attempt to understand the evolution that creates their great diversity.
References
- Alexandre H (2020) Wine yeast terroir: Separating the wheat from the chaff-for an open debate. Microorganisms 8(5): 787.
- Combina M, Elía A, Mercado L, Catania C, Ganga A, et al. (2005) Dynamics of indigenous yeast populations during spontaneous fermentation of wines from Mendoza, Argentina. International Journal of Food Microbiology 99 (3): 237-243.
- Raspor P, Milek MD, Polanc J, Mozina SS, Cadez N (2006) Yeasts isolated from three varieties of grapes cultivated in different locations of the Dolenjska vine-growing region, Slovenia. International Journal of Food Microbiology 109(1-2): 97-102.
- Schuller D, Valero E, Dequin S, Casal M (2004) Survey of molecular methods for the typing of wine yeast strains. FEMS Microbiology Letters 231 (1): 19-26.
- Martins G, Sertier MC, Lauga B, Claisse O, Funel LA, et al. (2012) Grape berry bacterial microbiota: Impact of the ripening process and the farming system. Int J Food Microbiol 158(2): 93-100.
- Ranjard L, Echairi A, Nowak V, Lejon DPH, Nouaïm R, et al. (2006) Field and microcosm experiments to evaluate the effects of agricultural Cu treatment on the density and genetic structure of microbial communities in two different soils. FEMS Microbiol Ecol 58(2): 303-315.
- Martins G, Vallance J, Mercier A, Albertin W, Stamatopoulos P, et al. (2014) Influence of the farming system on the epiphytic yeasts and yeast-like fungi colonizing grape berries during the ripening process. Int J Food Microbiol 177: 21-28.
- Patrignani F, Montanari C, Serrazanetti DI, Braschi G, Vernocchi P, et al. (2017) Characterisation of yeast microbiota, chemical and sensory properties of organic and biodynamic Sangiovese red wines. Ann Microbiol, pp.99-109.
- Bisson FL, Carpel JE (2010) Genetics of yeast impacting wine quality. Annu Rev Food Sci Technol 1: 139-162.
- Llaubères CRM (1993) Enzymes in winemaking. In: Fleet GH (Ed.), Wine Microbiology and Biotechnology. Harwood Academic Publishers, UK, pp. 477-506.
- Caridi, A, Cufari JA, Ramondino D (2002) Isolation and clonal pre-selection of enological saccharomyces. The Journal of General and Applied Microbiology 48(5): 261-267.
- Chalvantzi I, Banilas G, Tassou C, Nisiotou A (2021) Biogeographical regionalization of wine yeast communities in Greece and environmental drivers of species distribution at a local scale. Frontiers in Microbiology 12: 705001.
- Vergeade J, Guiraud J, Larpent LP (1976) Study of saint nectarine cheese yeasts. Lait 56: 555-556.
- Lodder J (1970) The yeasts. A taxonomic study. überarbeitete Auflage. X und 1385 S., 354 Abb. Amsterdam-London 1970: North Holland Publ. Co.
- Koehler AP, Chu KC, Houang ETS, Cheng AFB (1999) Simple, reliable, and cost-effective yeast identification scheme for the clinical laboratory. J Clin Microbiol 37(2): 422-426.
- White TJ, Bruns T, Lee S, Taylor (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Shinsky JJ, White TJ (Eds.), PCR protocols: A guide to methods and applications, Academic Press, Inc., New York, USA, pp. 315-322.
- Legras JL, Karst F (2003) Optimisation of interdelta analysis for Saccharomyces cerevisiae strain characterisation. FEMS Microbiol Lett 221(2): 249-255.
- Nadal D, Colomber B, Piña B (1996) Molecular polymorphism distribution in phenotypically distinct populations of wine yeast strains. Appl Environ Microbiol 62(6): 1944-1950.
- Suranská H, Vránová D, Omelková J (2016) Isolation, identification and characterization of regional indigenous Saccharomyces cerevisiae strains. Brazilian journal of microbiology 47(1): 181-190.
- Strauss MLA, Jolly NP, Lambrechts MG, Rensburg P (2001) Screening for the production of extracellular hydrolytic enzymes by non-saccharomyces wine yeasts. Journal of Applied Microbiology 91(1): 182-190.
- Comitini F, Gobbi M, Domizio P, Romani C, Lencioni L, et al. (2011) Selected non-Saccharomyces wine yeasts in controlled multistarter fermentations with Saccharomyces cerevisiae. Food Microbiology 28(5): 873-882.
- Parish ME, Caroll DE (1985) Indigenous yeasts associated with muscadine Vitis rotundifolia grapes and musts. American Journal of Enology and Viticulture 36(2): 165-169.
- Jolly NP, Varela C, Pretorius IS (2014) Not your ordinary yeast: Non-Saccharomyces yeasts in wine production uncovered. FEMS Yeast Research 14(2): 215-237.
- Zambonelli C (1998) Wine microbiology and biotechnology. Bologna, p. 300.
- Rupela OP, Tauro P (1984) Isolation and characterization of low hydrogen sulfide producing wine yeast. Enz Microbiol Techn 6: 419-421.
- Rodriguez CL, Joseph CML, Nazaris B, Coulon J, Richardson S, et al. (2020) Innovative use of non-saccharomyces in bio-protection: T. Delbrueckii and M. Pulcherrima applied to a machine harvester. Catalyst Discovery into Practice 4: 82-90.
- Bisson LF, Kunkee RE (1991) Microbial interactions during wine production. Mixed cultures in biotechnology. In: Zeikus JG, Johnson EA (Eds.), McGraw-Hill, New York, USA, pp. 39-68.
- Frezier V, Dubourdieu D, (1992) Ecology of yeast strain Saccharomyces cerevisiae during spontaneous fermentation in a Bordeaux winery. Am J Enol Vitic 43: 375-380.
- Granchi L, Ganucci D, Messini A, Rosellini D, Vicenzini M, at al. (1998) Dynamics of yeast populations during the early stages of natural fermentations for the production of Brunello de Montalcino wines. Food Technol Biotechnol 36: 313-318.
- Fleet GH (2003) Yeast interactions and wine Flavour. International Journal of Food Microbiology 86(1-2): 11-22.
- Nisiotou Α, Spiropoulos ΑΕ, Nychas GIE (2007) Yeast community structures and dynamics in healthy and botrytis-affected grape must fermentations. Applied And Environmental Microbiology 13(21): 6705-6713.
- Banilas G, Sgouros G, Nisiotou A (2016) Development of microsatellite markers for Lachancea thermotolerans typing and population structure of wine-associated isolates. Microbiological Research 193: 1-10.
- Mills DA, Johannsen EA, Cocolin L (2002) Yeast diversity and persistence in Botrytis-affected wine fermentations. Appl Environ Microbiol 68(10): 4884-4893.
- Valero E, Cambon B, Schuller D, Casal M, Dequin S (2007) Biodiversity of saccharomyces yeast strains from grape berries of wine-producing areas using starter commercial yeasts. FEMS Yeast Research 7(2): 317-329.
- Nikolaou E, Soufleros EH, Bouloumpasi E, Tzanetakis N (2006) Selection of indigenous saccharomyces cerevisiae strains according to their oenological characteristics and vinification results. Food Microbiol 23(2): 205-211.
- Tofaloa R, Chaves LC, Di Fabioa F, Schironea M, Felis GE, et al. (2009) Molecular identification and osmotolerant profile of wine yeasts that ferment a high sugar grape must. International Journal of Food Microbiology Volume 130(3): 179-187.
- Sun Y, Li E, Qi X, Liu Y (2015) Changes of diversity and population of yeasts during the fermentations by pure and mixed inoculation of Saccharomyces cerevisiae Ann Microbiol 65: 911-919.
- Schütz M, Gafner J (1993) Analysis of yeast diversity during spontaneous and induced alcoholic fermentations. J Appl Bacteriol 75(6): 551-558.
- De La Torre MJ, Millan MC, Juan PP, Morales J, Ortega JM (1999) Indigenous yeasts associated with two Vitis vinifera grape varieties cultured in southern Spain. Microbios 100(395): 27-40.
- Blanco P, Avalos MJM, Orriols I (2012) Effect of must characteristics on the diversity of Saccharomyces strains and their prevalence in spontaneous fermentations. J Appl Microbiol 112(5): 936-944.
- (2001) Environmental indicators for agriculture. Organization for economic cooperation and development (OECD), France, volume 3.
- Stenberg B (1999) Monitoring soil quality of arable land: Microbiological indicators. Acta Agric Scand 48: 1-24.
- Buckley DH, Schmidt TM (2001) The structure of microbial communities in soil and the lasting impact of cultivation. Microb Ecol 42(1): 11-21.
- Johnsen K, Jacobsen CS, Torsvik V, Sorensen J (2001) Pesticide effects on bacterial diversity in agricultural soils-a review. Biol Fertil Soils 33: 443-453.
- Ernest SKM, Brown JH (2001) Homeostasis and compensation: The role of species and resources in ecosystem stability. Ecology 82(8): 2118-2132.
- Kremer A, Zanetto A, Ducousso A (1995) Multilocus and multitrait measures of differentiation for gene markers and phenotypic traits. Genetics 145(4): 1229-1241.
- Cordero BG, Arrayo T, Serrano A, Tello J, Aporta I, et al. (2011) Influence of the farming system and vine variety on yeast communities associated with grape berries. International Journal of Food Microbiology 145(1): 132-139.
- Milanovic V, Comitini F, Ciani M (2013) Grape berry yeast communities: Influence of fungicide treatments. Int J Food Microbiol 161(3): 240-246.
- Ilieva F, Petrov K, Velickovska SK, Gunova N, Dimovska V, et al. (2021) Influence of autochthonous and commercial yeast strains on fermentation and quality of wines produced from vranec and cabernet sauvignon grape varieties from tikveš wine-growing region, republic of north macedonia. Appl Sci 11(13): 6135.
- Ganucci D, Guerrini S, Mangani S, Vincenzini M, Granchi L (2018) Quantifying the effects of ethanol and temperature on the fitness advantage of predominant Saccharomyces cerevisiae strains occurring in spontaneous wine fermentations. Front Microbiol 9: 1563.
- Vezinhet F, Hallet JN, Valade M, Poulard A (1992) Ecological survey of wine strains by molecular methods of identification. Am J Enol Vitic 43: 83-86.
- Sabate J, Cano J, Querol A, Guillamon JM (1998) Diversity of Saccharomyces strains in wine fermentations: Analysis for two consecutive years. Lett Appl Microbiol 26(6): 452-455.
- Granchi L Ganucci D, Viti C, Giovannetti L, Vincenzini M (2003) Saccharomyces cerevisiae biodiversity in spontaneous commercial fermentations of grape musts with adequate and inadequate assimilable-nitrogen content. Lett Appl Microbiol 36(1): 54-58.
- Granchi L, Ganucci D, Buscioni G, Mangani S, Guerrini S (2019) The biodiversity of saccharomyces cerevisiae in spontaneous wine fermentation: The occurrence and persistence of winery-strains. Fermentation 5: 86.
- Pérez CV, Ayala F, Echávarri JR, Negueruela AI (2003) Proposal for a new standard OIV method for determination of chromatic characteristics of wine. Am J Enol Vitic 54: 59-62.
- Pretorius I (2000) Tailoring wine yeast for the new millennium: Novel approaches to the ancient art of winemaking. Yeast 16(8): 675-729.
© 2023 Aikaterini Karampatea. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






