- Submissions

Full Text
Journal of Biotechnology & Bioresearch
Rapid and Sensitive SARS-CoV-2 Detection in Saliva Samples; Basis for a High Throughput Screening
Carmen Almaraz1, Jose María Marimón2, Ane Sorarrain2, Olatz Arrizabalaga3, David Otaegui4 and Ana Gorostidi1*
1Biodonostia Health Research Institute, Genomic Platform, Spain
2Biodonostia Health Research Institute, Infectious Diseases Area, Group of Respiratory Infection and Antimicrobial Resistance, Osakidetza Basque Health Service, Donostia University Hospital, Microbiology Department, Spain
3Biodonostia Health Research Institute, Group of Innovation, Spain
4Biodonostia Health Research Institute, Neuroscience Area, Group of Multiple Sclerosis, Spain
*Corresponding author: Ana Gorostidi, Biodonostia Health Research Institute, Genomic Platform, Spain
Submission: February 02, 2022;Published: March 16, 2022

Volume3 Issue4March, 2022
Abstract
We used a commercially available colorimetric enzyme mix to perform Reverse Transcribed Loop- Mediated Isothermal Amplification (RT-LAMP) for detecting SARS-CoV-2 on saliva samples without a previous RNA purification step. We describe our optimization of the LAMP assay that enabled simple, fast and sensitive detection in any saliva sample. Adding Chelex-100 and SDS to saliva samples, we achieved 97% efficiency in the detection of SARS-Cov-2 with spike-in saliva samples and 100% in a small sample of saliva from infected patients. This method is meant to be the basis for high throughput screening, as well as a feasible and cheap method for any laboratory.
Keywords: Covid-19; SARS-CoV-2; RT-LAMP; Saliva; Direct RNA detection
Introduction
The whole word has been suffering the Covid-19 pandemic for over a year now, with terrible sanitary, economic and sociological consequences. Huge investment and research efforts have been done so far to develop and implement sensitive and specific detection assays in order to properly detect, trace and isolate disease focus.
Detection of SARS-CoV-2 RNA by Reverse-Transcription real time PCR (RT-qPCR) in extracted RNA from nasopharyngeal swabs has become the gold standard worldwide as diagnostic tool. This is the most specific and sensitive methodology, but a more economic and fast technology is needed for large screenings. With this in mind, we try to develop a SARSCoV- 2 detection method that will enable the screening of large populations in a short time and in an economic way. We believe that this strategy might help detecting disease focus and therefore, prevent its spread. In this race for better (cheaper and faster) tools to detect the SARSCoV- 2, LAMP technology, an alternative to conventional RT-qPCR, has been adjusted for SARSCoV- 2 diagnostic. In fact, previous works have intended to develop fast and sensitive assays using LAMP technology with different assays [1-4]. Loop-Mediated Isothermal Amplification (LAMP) is well known for its robust and highly sensitive and specific amplification of target nucleic acid, which is achieved by utilizing up to six primers at a constant temperature. It is a method developed by Eiken Chemical Co., Ltd. that has successfully been used before for diagnosis of infectious diseases such as tuberculosis, malaria, sleeping sickness, Ebola and Zika [5]. In a pandemic scenario, isothermal amplification is essential. because it allows the technique to be scaled up to high throughput.
On the other hand, saliva is a promising sample for testing due to ease, safe and noninvasive nature of its collection. Recently, saliva-based tests were approved by the FDA for SARS-CoV-2 routine diagnostics, as the saliva analysis was found to be more consistent than nasopharyngeal swabs in direct comparison (Wyllie et al., 2020). Moreover, it’s relatively high viral load and proved consistent and sensitive results support the use of saliva in high throughput screenings [6,7]. In this scenario, avoiding RNA extraction is mandatory. However, only a few works have tried the implementation of direct detection without a previous RNA extraction, and few of these works have been validated in clinic samples. In the present work, we develop a SARS-CoV-2 LAMP detection method on saliva sample with a direct, fast, sensitive and economic workflow that will allow large-scale screenings of population and might as well be a starting premise for future infectious disease screenings.
Results
We first searched for a suitable commercial LAMP assay for SARS-CoV-2 detection that would satisfy our necessities: one-step amplification (retro-transcription included), colorimetric assay, direct sample impute (without nucleotide purification needed) and saliva sample proved. None of the commercially available assays fulfilled all the criteria, but one was close: SARS-CoV-2 Rapid Colorimetric LAMP Assay Kit from New England Biolabs, USA. rActine gene as internal control, which will amplify the host’s genome in the sample, as well as the assay for SARS-CoV-2 with two targets (N1 and E gene), will turn from pink to yellow when positive amplification is achieved allowing a naked-eye interpretation of colorimetric LAMP.
We used spiked-in saliva samples for testing the assay. As spike, several SARS-CoV-2 RNA extracted from nasopharyngeal samples using the STARMag universal extraction system (Seegene Inc, Seoul, South Korea) with Ct between 15 and 20 for the E, N and RdRP genes as detected by RT-PCR using the Allplex 2019-nCoV assay (Seegene) were used. A dilution 1:10 of this RNA was used for the different experiments. The Positive Control (PC) of the commercial kit was also used as spike and in serial dilutions for sensibility testing. This PC is a target DNA for the SARS-CoV-2 assay with approximately 10⁶ copies/μl. All the incubations were performed in a thermocycler (30min at 65 ºC) in 0.2ml reaction tubes.
The basic protocol (NEBmanuale2019.pdf; Master Mix, Oligo mix, guanidine chloride, saliva sample, ddH2O), although successful with different isolated RNA, was not consistent with saliva samples and therefore, a range of compounds commonly used to enhance PCR or other isothermal methods was tested. Most of the additives aimed to buffer basal pH differences in saliva (TE, TBE, NaOH), inhibit DNA binding proteins or RNA degrading (proteinase K, Rnase inhibitor (RNase Out, Invitrogen), MycroLYSIS-RNA (Clent Life Science), Chelex-100 (Bio-Rad #1421253)) and/or enhance RNA amplification (DTT, SDS, Tween). In contrast to SARS-CoV-2 assay, the assay for rActine amplification does not present problems with direct saliva samples. One reason could be that SARS-CoV-2 assay is a mix of two assays and might have bigger limitations amplifying the virus targets.
Plenty of studies have come out using PBS, TE, water or NaOH to dilute saliva samples [2,8]. However, in our experience, SARS-CoV-2 was inconsistently detected in many saliva samples indicating the presence of an inhibitor in saliva that impaired the assay. Many of the mentioned works [9] have not tried with different saliva samples but with technical replicates and this could mitigate the putative effect of inhibitors. Previous works showed that a key step is the heating of the sample previous to the LAMP reaction [2,8]. It dramatically increases sensitivity of target amplification as it provides RNA release and nucleases inactivation in crude samples. When working with purified RNA, this step should not be necessary, however, as our protocol aims to scale up to infectious saliva, we decided to implement this step of virus inactivation/RNA extraction (95 ºC for 10min) from the beginning.
Lalli MA et al. [8] found that LAMP reaction is compatible with samples diluted in Phosphate Buffered Saline (PBS) or TE buffer (10mM Tris, 0.1mM EDTA), and that heat treatment (55 ºC for 15 minutes, 95 ºC for 5 minutes) with and without proteinase K improved viral detection compared to untreated samples (p<0.01 and p<0.001 respectively). However, neither protease treatment of saliva samples, nor the above-mentioned compounds helped to obtain the desirable results in our laboratory. Only Chelex-100 protocol showed good enough efficiency (71% as shown in four experiments with 23 different saliva samples). In keeping with standard Chelex-100 protocol, samples were mixed 2:3 with 30% wt/vol Chelex-100 solution, incubated at 95 ºC for 10 minutes, and then assayed with RT-LAMP. Pulse spinning pretreated saliva samples in a microfuge immediately prior addition to the LAMP reaction showed to be necessary. Given the good results observed with Chelex-100, we sought to improve sensitivity incorporating new compounds into our highest sensitivity protocol. The best results were archived when SDS at a 0.5% final concentration was added to the sample before Chelex, showing a 97% of efficiency. All experiments of RT-LAMP were run repeatedly and in triplicates to verify reproducibility.
Our best results were obtained when saliva samples were mixed with Chelex. This resin has shown to be crucial and much more effective than another buffer, enzyme or alkalinizer. By sequestering divalent metal cations, Chelex-100 protects RNA from degradation at high temperature. Chelex-100 also has a basic pH, which reduces sensitivity to variations in patient saliva pH without inhibiting LAMP. We observed that mixed with SDS, results were better as samples that would not amplify with only Chelex would work as well when SDS was added to the mix. In this sense, it is worth noting that unlike described before by other authors, the addition of guanidine chloride to LAMP reaction, significantly improves the percentage of positive detection, increases performance and was fundamental for the SARS-CoV-2 LAMP assay in our experiments, although it was needless for the rActine assay.
SARS-Cov-2 infected RNA or RNA containing the LAMP assay target was spiked into negative control saliva samples to simulate a virus-infected saliva. This is a useful model that allows testing different saliva types with no need to manipulate virus-infected saliva in the laboratory. However, and to verify that our assay and protocol have the same efficiency with infected saliva, we tested 9 saliva samples available from de Microbiology department of the Hospital Donostia. In this case, all of the saliva’s positive for SARS-Cov-2 were detected as positive as well with LAMP assay. 20 negative saliva were included in the experiment as negative control and were tested as well after spiked-in with SARS-Cov-2 RNA (Figure 1). The results confirm that none of the saliva samples inhibited the assay reaction and 100% of the positive samples were diagnosed as positive.
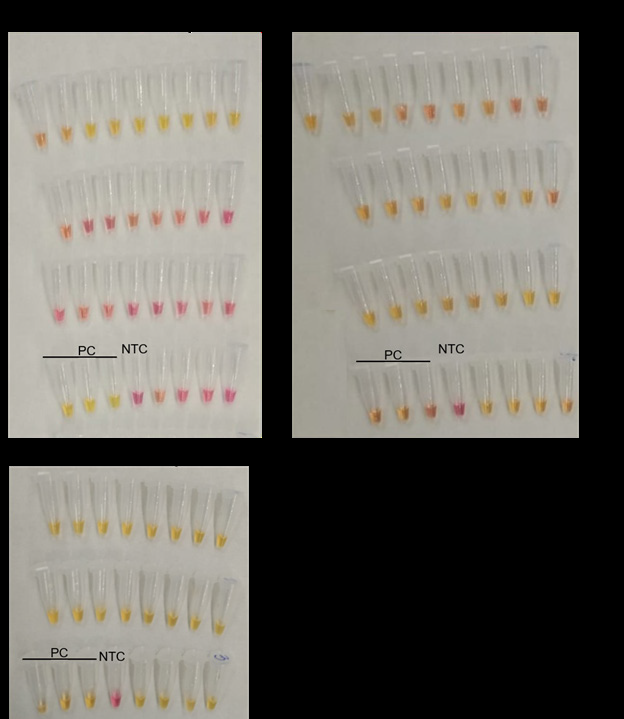
Figure 1: Analysis of SARS-Cov-2 infected saliva samples with LAMP assays. Yellow colour denotes positive amplification of the reaction whereas pink indicates no amplification.
A. Virus infected salivas, positive for SARS-Cov-2 with RT-qPCR assays, were detected positive as well with LAMP assay. 20 virus free saliva were included in the experiment as negative controls.
B. The same samples (9 infected saliva and 20 virus free saliva) were tested for rActine assay.
C. The same 20 SARS-Cov-2 negative saliva samples were tested after spiked-in with SARS-Cov-2 RNA to show that none of the saliva inhibited SARS-Cov-2 LAMP assay.
PC: positive control for SARS-Cov-2.
NTC: None Template Control.
We tested the SARS-CoV-2 assay for specificity. Various respiratory virus RNA samples were checked (Influenza A H1 andH3, Influenza B, RSV, Bocavirus, Rhinovirus) and for all of them the result was negative for SARS-CoV-2. Finally, we determined de Limit of Detection (LOD) of our optimized assay using serial dilutions of the virus positive control template (NEB) spiked into saliva. The assay detects a minimum of 20 total RNA copies of SARSCoV- 2 per reaction (10 copies/μl. of sample in the reaction). Since saliva samples were 5-fold diluted (because SDS and Chelex are added) prior to LAMP reaction, the detection limit of the assay with saliva is above 50 copies/μl.
The colorimetric detection in this method is based on the pH-sensitive dye phenol red. Incorporation of dNTPs during DNA amplification results in accumulation of protons that reduces the pH in the reaction buffer and can be detected by a color change. In acidic pH (pH≤6.8), phenol red changes color to yellow. The simplicity of the assay and workflow makes it useful for any house made laboratory without the necessity of sophisticated equipment. However, fluorophore-based assays give a more precise interpretation of the results as well as a quantification of target copies [10].
Conclusion
We conclude that an efficient and sensitive detection of SARSCoV- 2 in saliva can be achieved by colorimetric RT-LAMP with SDS and Chelex addition to the sample, a short spin and a 10min incubation at 95 ºC prior to LAMP amplification at 65 ºC for 30min. Our approach solves two major bottlenecks in massively screening for COVID-19 causing infectious virus: sample collection and RNA extraction. However, future studies are needed with higher number of saliva samples from SARS-CoV-2 infected patients at different timepoints of the infection to confirm the test as a high throughput screening diagnostic method with sufficient sensitivity.
References
- Yu L, Wu S, Hao X, Li X, Liu X, et al. (2020) Rapid colorimetric detection of COVID-19 coronavirus using a Reverse Transcriptional Loop-Mediated Isothermal Amplification (RT-LAMP) diagnostic platform. Clin Chem 6(7): 975-977.
- Yang Q, Meyerson NR, Clark SK, Paige CL, Fattor WT, et al. (2021) Saliva twostep for rapid detection of asymptomatic sars-cov-2 carriers. medRxiv.
- Zhang Y, Ren G, Buss J, Barry AJ, Patton GC, et al. (2020) Enhancing colorimetric loop-mediated isothermal amplification speed and sensitivity with guanidine chloride. Biotechniques 69(3):179-185.
- Reynés B, Serra F, Palou A (2021) Rapid visual detection of SARS-CoV-2 by colorimetric loop-mediated isothermal amplification. Biotechniques 70(4): 218-225.
- Bhadra S, Riedel TE, Lakhotia S, Tran ND, Ellington AD (2021) High-surety isothermal amplification and detection of SARS-Cov-2. mSphere 6(3): e00911- e00920.
- Nagura-Ikeda M, Imai K, Tabata S, Miyoshi K, Murahara N, et al. (2020) Clinical evaluation of self-collected saliva by quantitative reverse transcription-PCR (RT-qPCR), direct RT-qPCR, reverse transcription-loop-mediated isothermal amplification, and a rapid antigen test to diagnose COVID-19. J Clin Microbiol 58(9): 1-9.
- Taki K, Yokota I, Fukumoto T, Iwasaki S, Fujisawa S, et al. (221) SARS-CoV-2 detection by fluorescence loop-mediated isothermal amplification with and without RNA extraction. J Infect Chemother 27(2): 410-412.
- Lalli MA, Langmade SJ, Chen X, Fronick CC, Sawyer CS, et al. (2021) Rapid and extraction-free detection of SARS-CoV-2 from saliva with colorimetric LAMP. Clin Chem 67(2): 415-424.
- Howson ELA, Kidd SP, Armson B, Goring A, Sawyer J, et al. (2021) Preliminary optimisation of a simplified sample preparation method to permit direct detection of SARS-CoV-2 within saliva samples using Reverse-Transcription Loop-Mediated Isothermal Amplification (RT-LAMP). J Virol Methods 289: 114048.
- Mautner L, Baillie CK, Herold HM, Volkwein W, Guertler P, et al. (2020) Rapid point-of-care detection of SARS-CoV-2 using Reverse Transcription Loop-Mediated Isothermal Amplification (RT-LAMP). Virol J 17(1): 1-14.
© 2022 Ana Gorostidi. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






