- Submissions

Full Text
Journal of Biotechnology & Bioresearch
The assembly formation by co-cultures of DNA (Escherichia coli) crown cells with Bacillus subtilis cells
Shoshi Inooka*
Japan Association of Science Specialists, Japan
*Corresponding author: Shoshi Inooka, Japan Association of Science Specialists, Japan
Submission: April 9, 2021;Published: May 3, 2021

Volume2 Issue5May, 2021
Abstract
DNA crown cells are artificial cells in which the outside of the membrane is covered with DNA. Such artificial cells are generated readily using sphingosine (SPH)-DNA-adenosine mixtures and can proliferate within egg whites. To date, many kinds of DNA crown cells have been synthesized using DNA from various donors.
During previous studies of DNA crown cells, it was observed that antibiotics are produced by the coculture of DNA crown cells with yeast (from beer). Based on these findings, I inferred that DNA crown cells may control the growth of yeast and that such DNA crown cells may multiply. In initial experiments to clarify such problems, Bacillus subtilis (natto), an organism that presents several advantages. were employed instead of yeast.
In the experiments described here, it was demonstrated that assemblies of many sizes and shapes are formed in the mixtures of DNA (Escherichia coli) crown cells synthesized using adenosine-monolaurin compounds with B. subtilis cells. Here, characteristic assemblies were observed to be generated from such mixtures. In some assemblies, objects resembling DNA crown cells and bacteria were observed, suggesting that such cells may be incorporated into these assemblies, with associated suppression of bacterial growth.
Keywords: DNA crown cells; Assembly; Bacillus subtilis; Sphingosine-DNA
Introduction
There has been significant progress in the generation of artificial cells. Recently, approaches for generating fully operational (self-replicating) artificial cells have been reported [1,2]. Such artificial cells (which have been designated DNA crown cells) [3] are covered with DNA and are generated upon incubation with egg white. In theory, unlimited numbers of DNA crown cells could be prepared using the described methods and several kinds of DNA crown cells have been generated [4-7].
On the other hand, it is not clear whether such DNA crown cells have useful biological functions, notably including possible contributions in the field of applied science. In previous experiments [8,9], it was reported that antibiotics are produced in co-cultures of DNA crown cells generating with DNA from Streptomyces griseus, an actinomycete that produces several kinds of antibiotics with yeast (from beer).
In the course of a study intended to clarify the mechanism of antibiotic production by these DNA crown cells, it was demonstrated that the DNA crown cells suppressed growth of the yeast that were co-cultured with the DNA crown cells, suggesting that the DNA crown cells may inhibit the yeast metabolism or propagation [10]. In further experiments, we sought to clarify whether DNA crown cells do indeed inhibit the growth of yeast, and whether the DNA crown cells were themselves able to grow.
However, these tests were preliminary experiments. We found that there were challenges to the use of yeast in these studies. Notably, it was difficult to distinguish between yeast and DNA crown cells solely based on light microscopy, although the characteristic shape of yeast (oval) was distinct from that of DNA crown cells (round). Therefore, for the first set of experiments seeking to clarify whether DNA crown cells influenced the growth of yeast or bacteria, we employed Bacillus subtilis, a rod-shaped bacterium, instead of yeast. DNA crown cells were generated using SPH, DNA (E. coli) and adenosine –monolaurin compounds. Assemblies were observed as soon as such DNA crown cells were mixed with B. subtilis suspended in water. Multiple types of assemblies (varying in sizes and shapes) were formed.
Given the variations in these assemblies, we first sought to define the appearance of these assemblies. For examples, when the mixtures of DNA crown cells combined with B. subtilis were incubated for 3 hours at 37 ºC, many bacteria-like objects or many objects that were similar to DNA crown cells were observed within the assemblies, suggesting that the B. subtilis cells had been enclosed within the assemblies and DNA crown cells may be incorporated into or generated within these assemblies.
In the present experiments, we observed that many kinds of the assemblies that varied in size and shape were formed in the mixtures of DNA crown cells with B. subtilis. Typically, bacteria appeared to be enclosed within the assemblies, and several assemblies possessed multiple objects that resembled DNA crown cells. However, to date, it remains unclear whether these objects are DNA crown cells or B subtilis. Nonetheless, I did observe that the growth of B. subtilis in the co-cultures was strongly suppressed (data not shown), a point that I will address elsewhere.
On the other hand, it is unclear whether DNA crown cells multiply in the co-cultures, because I could not prove that the round objects that resembled DNA crown cells actually were DNA crown cells. Thus, the ability of DNA crown cells to propagate remains unclear. This ambiguity suggests a possible new function of DNA crown cells, such that some objects may be generated from the assemblies that form using materials from DNA crown cells as sources of new cells. Such experimental findings; implying the generation of new objects from the assembly, may contribute to the development of multiple aspects of the fields of basic and applied life sciences.
Materials and Methods
Materials
The following materials were used: SPH (Sigma, USA and Tokyo Kasei, Japan), DNA (E. coli, B1 strain; Sigma-Aldrich, USA), adenosine (Sigma, USA and Wako, Japan), monolaurin (Tokyo Kasei, Japan) A-M compound (synthesized from a mixture of adenosine and monolaurin) [8], The bacterial culture medium consisted of Sanisupetsuku growth medium (Azuwan, Japan). B. subtilis natto (B. subtilis) was obtained as Dry Bacillus subtilis (Daikokuya, Nagoya, Japan). Actively growing (cultured) B. subtilis cells were used. In initial experiments, dry B. subtilis was suspended in distilled water at approximately 1.0mg per 1.5mL. In subsequent experiments, a portion of dry B. subtilis was cultivated on solid medium (plates) at 37 ºC for 18 hours using bacterial culture medium. The cultured bacteria were harvested and re-suspended in distilled water to a density of approximately 106/mL.
Methods
Preparation of DNA crown cells: The generation of artificial cells using SPH-DNA-A-M was performed as described previously [8]. Briefly, l80μg of SPH (10mM) and 90mL of DNA (1.7mg/mL) were combined, and the mixture was heated twice. A-M compound (100mL) was added, and the mixture then was incubated at 37 ºC for 15 minutes. Next, 30μL of monolaurin was added, and the mixture was incubated at 37 ºC for another 5 minutes. The resulting suspension was used as the inoculum of DNA crown cells. The mixture of B. subtilis with DNA crown cells: First, 50μL of the B. subtilis suspension (prepared as described above) was combined with 50μL of DNA crown cells. A portion of the mixture (prepared at room temperature) was observed immediately. The remainder of the mixture was incubated for 3 hours at 37 ºC, at which point a sample again was observed.
Microscopic observations: A drop of the DNA crown cells that were cultured with B. subtilis were placed on a slide glass and topped with a cover glass. The resulting slide then was observed under microscopy. In summary, the samples observed were as follows:
A. The samples immediately after mixing.
B. The samples after incubation for 3 hours at 37 ºC.
Results and Discussion
Microscopic appearance of the combination of DNA crown cells and B. subtilis
Microscopic appearance of DNA crown cells: DNA crown cells were prepared using commercially purchased E. coli DNA combined with adenosine-monolaurin.
A typical single DNA crown cell is shown in Figure 1 (arrow a). A typical single DNA crown cell is shown here (Figure 1, arrow a). DNA crown cells are usually round in shape and may be surrounded by a cloud-like layer. Such cells are usually approximately 2-3μm in diameter (Figure 2, arrows b and c). A typical round-shaped cell is also observed in this image (Figure 1, arrow c). It is approximately 10μm in size, and is shown in Figure 2 (arrows a and d). Clusters of DNA crown cells can be observed (Figure 1, arrow b). Objects other than DNA crown cells are also seen (Figure 1, arrow d).These cells are round and sometimes are surrounded by a cloud-like layer. These cells typically are 2-3μm in diameter (Figure 2, arrows b and c), as indicated by the objective micrometer, in which the length of one interval is 10μm. Other round cells of various sizes also were observed (Figure 1, arrow c). A typical cell of the latter type is indicated by arrows a and d in Figure 2 one division of the micrometer indicates a length of 10μm. A single DNA crown cell is indicated by arrows b and c. Both of these objects have sizes of approximately 2-3μm in diameter. Two other round DNA crown cells are indicated by arrows a and d; both of these objects have diameters of approximately 10μm. this cell was approximately 10μm in size. Clusters that consisted of several DNA crown cells also were observed (Figure 1, arrow b). Objects other than cells were also seen (Figure 1, arrow d). Such objects may be assemblies of the component materials (i.e., SPH, DNA, SPH-DNA, and A-M compound) that were not incorporated into the DNA crown cells. The objects had several different colors, but no consistent color identification of specific objects was observed. Nonetheless, the color of the objects under microscopy did facilitate the observation of the targets.
Figure 1:Microscopic appearance of DNA crown cells.
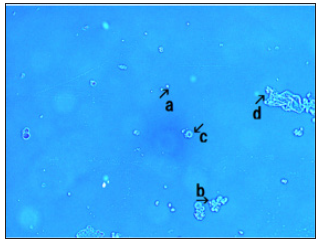
Figure 2: DNA crown cells under the micrometer.
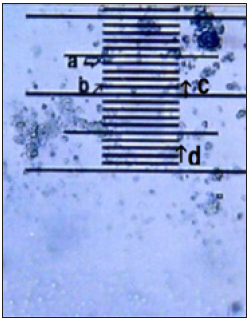
Figure 3: Microscopic observation of Bacillus subtilis.
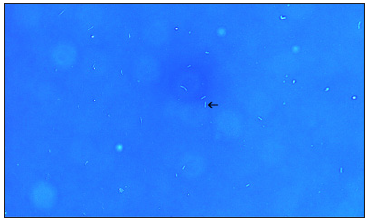
Figure 4:This image shows clustering of several fibrous regions with DNA crown cells (Figure 4, arrow a). A free DNA crown cell also is observed (Figure 4, arrow b).
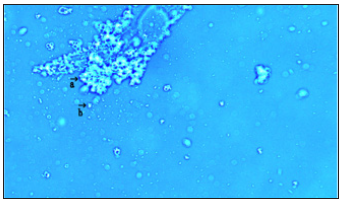
Figure 5:This image shows clustering of round DNA crown cells (Figure 5, arrow a). A free DNA crown cells also are observed (Figure 5, arrows b and c).
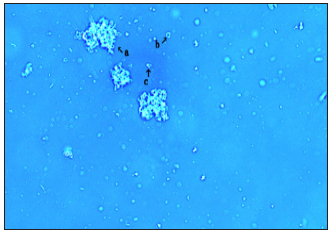
Figure 6: This image (Figure 6, arrow a) shows the clustering of several fibrous regions longer than those seen in Figure 4 & Figure 5.

Figure 7: This image (Figure 7) shows an assembly that is flat in appearance and contains objects resembling bacteria (Figure 7, arrow a) and round DNA crown cell-like objects (Figure 7, arrow b).
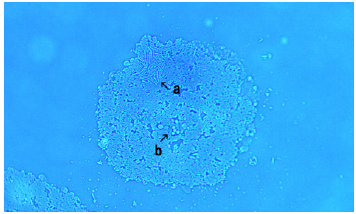
Figure 8: This image (Figure 8) shows an assembly that contains objects resembling bacteria (Figure 8, arrows a and b). A free bacteria also observed (Figure 8, arrow c).
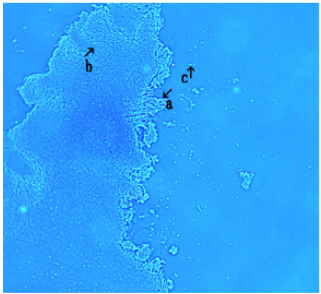
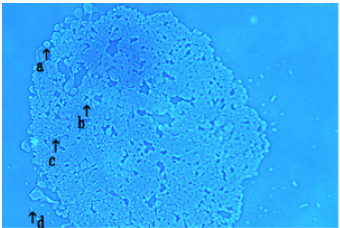
Figure 9: This image (Figure 9) shows an assembly that appears to possess round objects along its edge (Figure 9, arrow a ) and within assembly (Figure 9, arrow b). Also, it contains objects resembling bacteria (Figure 9 arrow c). A free bacteria is observed (Figure 9, arrow d).
Microscopic observation of B. subtilis: The B. subtilis used in these experiments are shown in Figure 3. A typical cell is indicated by arrow. This cell has a length of approximately 3~4μm. A typical cell (Figure 3 arrow) was rod-shape and approximately 3-4μm in length.
The formation of assemblies in mixtures of DNA crown cells with B. subtilis before incubation
When B. subtilis was combined with DNA crown cells and observed immediately, several kinds of assemblies (varying in size and shape) were observed. Here, several assemblies that may be characteristic are described. An assembly that appeared to consist of fibrous material was observed with several DNA crown cells, as shown in Figure 4. arrow a. The sizes of the assemblies are not shown in the present experiments, because the sizes of these assemblies may not be important. However, single DNA crown cells or single bacterial cells are shown in the figures when such cells were positioned in proximity to the assemblies. Given that DNA crown cells or bacteria are typically 2-4μm in size, the sizes of assemblies can be estimated by comparison. Other assemblies of similar types were also observed (Figures 5 & 6). The assembly in Figure 5 appears to consist of round DNA crown cells.
On the other hand, the differences between the assemblies may reflect differences in the sizes of the fibrous segment. Notably, the assembly in Figure 6 (arrow a) appears to consist of thicker and longer fibrous material than that seen in other assemblies. Several DNA crown cells also appear to be contained within the assembly pictured in Figure 6 (arrow b) Also, several DNA crown cells are observed within the assembly (Figure 6, arrow b). A free DNA crown cell also is observed (Figure 6, arrow c).
Microscopic appearance after 3 hours of incubation
After the incubation, many kinds of assemblies were observed. Characteristic post-incubation assemblies are shown in Figures 7-9. These assemblies were typically round in shape, as shown in Figure 7. Several objects were observed within such assemblies.
Objects
A. Shaped like bacteria were observed, as shown in Figure 7 (arrow a). Also, another type of objects
B. That were round in shape was observed (Figure 7 arrow
b). Other assemblies of types similar to those in Figure 7 were observed. As shown in Figure 8, the assembly appeared to possess many the bacteria-shaped objects seen in Figure 8. arrows a and b.
On the other hand, small round objects were observed along the edge of the assembly (Figure 9, arrow a). Microscopic observations suggested that these objects were also generalized within the assembly Figure 9 arrow.
Anyway, it is clear that these objects were generated from components of the assembly
Regarding the formation of these assemblies, it is clear that their formation had to have been based on the interactions between SPH and DNA. As previously reported [11,12], the long fibrous material is formed when SPH mixes with DNA, and DNA crown cells then are formed based on such fibrous assemblies. Therefore, the present assemblies may consist of DNA crown cells and other components that were not employed in the construction of DNA crown cells, as shown in Figure 1.
On the other hand, SPH or SPH-DNA is known to bind various compounds and to form various objects. Therefore, it is clear that the formation of the present assemblies was based on the binding of SPH-DNA to some compounds; which may influence the shape or size of the resulting assemblies. Above all, in the present experiments, it was demonstrated that assemblies were formed, and that objects resembling bacteria and DNA crown cells were observed within these assemblies or along their edges, suggesting that both classes of objects may be incorporated into the observed assemblies.
To date, it remains unclear whether the objects that resemble bacteria are indeed B. subtilis, or whether the objects that resemble DNA crown cells are in fact DNA crown cells. Nonetheless, it is clear that some objects were incorporated into the assemblies that were generated in these experiments, which included B. subtilis cells and DNA (E. coli) crown cells.
In this case, it is clear that the growth of B. subtilis was suppressed (data not shown), a result that will be addressed elsewhere. Hence, the same growth inhibitory effects seen previously with yeast appear to apply to the B. subtilis included in our experimental mixture. Further experiments will be needed to investigate whether such assemblies are formed in combinations of yeast and DNA (Streptomyces) crown cells, and whether DNA crown cells multiply. Such experiments are planned and are expected to clarify further the mechanisms of antibiotic production in cocultures of yeast and DNA crown cells [10].
The present study was the result of a new investigation of whether objects, including DNA crown cells and B. subtilis cells, were incorporated into the assemblies that are formed from SPHDNA mixtures. To clarify whether new objects were incorporated into these assemblies, and whether DNA crown cells generate new objects, it will be important to prepare various assemblies and to clarify their characteristics. To this end, further experiments are planned to test whether the assemblies that were observed here were formed from the combination of DNA crown cells, as opposed to DNA (E. coli) crown cells, with suitable bacteria, including B. subtilis.
References
- Inooka S (2012) Preparation and cultivation of artificial cells. App Cell Biol 25: 13-18.
- Inooka S (2016) Preparation of artificial cells using eggs with sphingosine-DNA. J Chem Eng Process Technol 7(1): 1-5.
- Inooka S (2016) Aggregation of sphingosine-DNA and cell construction using components from egg white. Integrative Molecular Medicine 3(6): 1-5.
- Inooka S (2017) Systematic preparation of bovine meat DNA crown cells app. Cell Biol, Japan 30: 13-16.
- Inooka S (2018) Systematic preparation of DNA (Akoya pearl oyster) crown cells. App Cell Biol, Japan 31: 21-34.
- Inooka S (2013) Preparation of artificial cells for yogurt production. App Cell Biol 26: 13-17.
- Inooka S (2014) Preparation of artificial human placental cells. App Cell Biol 27: 43-49.
- Inooka S (2019) Preparation of generated DNA (Streptomyces griseus) crown cells (Artificial cells) and antibiotic production in its’ co-cultures with yeast (Beer). Current Trends on Biotechnology & Microbiology 1(2): 26-32.
- Inooka S (2019) Antibiotic production in co-cultures of DNA (Stretomyces) crown cells (Artificial Cells) and yeast (Beer). Chemical & Pharmaceutical Research 1(2): 1-4.
- Inooka S (2020) On the mechanism of antibiotic production in co-cultures of generated DNA (Streptomyces Griseus) crown cells (Artificial cells) with Yeast (Beer). Novel Research in Science 5(3): 1-5.
- Inooka S (2000) Cytoorganisms (cell-original cultivable particles) with sphingosine-DNA. Communication in Applied Cell Biology 17(1-4): 11-34.
- Inooka S (2017) Biotechnical and systematic preparation of artificial cells (DNA crown cells). The Global Journal of Researches in Engineering 17(1-C): 1-10.
© 2021 Shoshi Inooka. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






