- Submissions

Full Text
Journal of Biotechnology & Bioresearch
Bioremediation of Petroleum Contaminated Environments by Pseudomonas Species
Stancu Mihaela Marilena
Institute of Biology Bucharest of Romanian Academy, Romania
*Corresponding author:Stancu MM, Institute of Biology Bucharest of Romanian Academy, 296 Splaiul Independentei, Romania
Submission: July 01, 2019;Published: June 10, 2019

Volume2 Issue1July, 2019
Abstract
The genus Pseudomonas is one of the most diverse and ubiquitous bacterial genera whose species was isolated from all types of environments, including from petroleum polluted environments. The genus Pseudomonas has significant biotechnological importance and comprises metabolically versatile bacteria capable to tolerate and degrade toxic petroleum hydrocarbons (e.g., aliphatic, aromatics). Several Pseudomonas strains are remarkable producers of secondary metabolites and enzymes which permit them to tolerate and degrade toxic petroleum hydrocarbons.
Keywords: Pseudomonas; Hydrocarbons; Tolerance; Degradation
Introduction
The genus Pseudomonas described formerly by Migula in 1894, is one of the most diverse and ubiquitous bacterial genera whose species was isolated worldwide from all types of environments, including from terrestrial, aquatic, human, animal, and plant host-associated environments [1-5]. The taxonomy of the genus Pseudomonas was disputed during the last decades. A lot of bacteria formerly included in Pseudomonas genus were reclassified in other genera or species, from the class Alpha-, Beta- or Gamma proteobacteria [1,2]. Currently, the genus Pseudomonas (sensu stricto) encompasses only species which belong to Pseudomonas RNA homology group I, from the class Gammaproteobacterial [2,4]. Two main phylogenetic branches or lineages, Pseudomonas aeruginosa and Pseudomonas fluorescens were discriminated in the genus Pseudomonas based on Multi-Locus Sequence Analysis (MLSA) of four housekeeping genes (i.e., 16S rRNA, gyrB, rpoB, rpoD) [2]. The number of the species in this genus is increasing every year [2,6]. The genus Pseudomonas has significant scientific and technological importance and comprises metabolically versatile bacteria capable to utilize a wide range of simple and complex organic compounds, as sources of carbon and energy [3,5,7]. Several Pseudomonas strains produce extracellular secondary metabolites (e.g., surfactants, pigments) [5,8] and extracellular enzymes (e.g., solvent-stable protease, lipase) [9,10], which permit them to tolerate and degrade simple and complex toxic organic compounds, including petroleum hydrocarbons [3,5].
Petroleum hydrocarbons, such as aliphatic and aromatics, are very toxic for most of the bacteria, because these molecules partition preferentially into the lipid bilayer of the cytoplasmic membrane, causing significant changes in their structure and fluidity [11-16]. These changes affect the vital functions of the cytoplasmic membrane, as selective permeability barrier, protein and reaction matrix, and as energy transducer, leading to loss of ions and intracellular macromolecules (e.g., RNA, proteins), changes in the electric potential and intracellular pH, inhibition of the bacterial cell metabolism and growth, and ultimately cell death [11,12,17]. The toxicity of hydrocarbons correlates with the logarithm of its partition coefficient in a standard octanol and water mixture (log Pow). Aliphatic and aromatics with a log Pow value between 1 and 5 are commonly toxic for bacteria even at very low concentrations. However, a number of bacteria including several Pseudomonas sp. strains are able to tolerate high concentrations of toxic hydrocarbons [3,11-15,17]. The toxicity of hydrocarbons for bacteria is correlated not only with its hydrophobicity expressed as log Pow, but also with the molecular structure of hydrocarbons and bacterial cell membranes composition [11,14,18]. The mechanisms of hydrocarbon tolerance in bacteria are not fully understood [11]. However, several possible adaptive mechanisms were described for bacteria able to survive and grow in the presence of toxic hydrocarbons:
1) changes of cells morphology (e.g., cells size increase or decrease).
2) changes in the membrane’s lipid bilayer (e.g., cis-trans isomerization of unsaturated fatty acids, saturation of fatty acids, phospholipid headgroups shift to reduce their fluidity and permeability, formation of membrane vesicles to transport hydrocarbons away from the cells).
3) active extrusion of the excess hydrocarbons from the cells to the outer medium by efflux systems (e.g., RND efflux pumps, ABC-efflux transporters).
4) activation of general stress response system (e.g., induction of DNA repair systems, activation of chaperons that stabilize and refold proteins, activation of oxidative stress proteins that remove reactive oxygen species).
5) enhanced energy production (e.g., induction of TCA cycle encoding enzymes, higher metabolism rate) [11-13,15-17,19].
Therefore, the tolerance of bacteria to hydrocarbons is the result of a multifactorial process, which involves a wide range of physiological and genetic changes which acts jointly for a complete adaptation [13,15]. There is no physiological correlation between the tolerance of bacteria to hydrocarbons and their degradation capability. A numberof Pseudomonas sp. strains were able to survive and grow in the presence of toxic hydrocarbons, but they were not able to degrade or modify them [12,15,17]. The Pseudomonas strains which possess specific enzyme systems (e.g., oxygenase’s and peroxidases, peripheral degradation enzymes) are able to degrade toxic hydrocarbons [20,21]. However, degradation may facilitate the resistance of bacterial cells to hydrocarbons, it cannot be the main mechanism involved in the hydrocarbon’s tolerance [12,15,17,20].
Conclusion
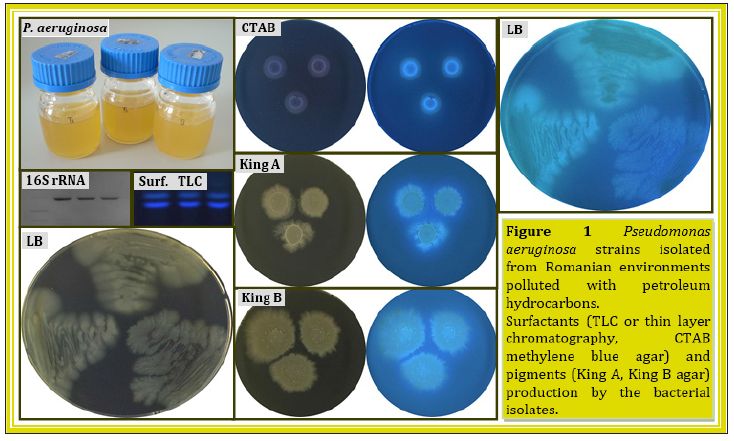
A number of bacteria able to tolerate and degrade toxic hydrocarbons were isolated from petroleum-rich environments, such as petroleum reservoirs and petroleum polluted sites. The abundance of hydrocarbon-degrading bacteria in these environments is correlated to the types of petroleum hydrocarbons and to the environmental factors [22]. Several Pseudomonas aeruginosa strains were isolated by us from various Romanian environments polluted with petroleum hydrocarbons [23,24]. The isolated Pseudomonas aeruginosa strains were able to survive and grow in the presence of hydrocarbons, including aliphatic and aromatics with a log Pow value between 2 to 5. The production of extracellular secondary metabolites, such as surfactants and pigments (Figure 1), was also observed for these bacteria. Like other Pseudomonas strains, these bacteria or their metabolites could have multiple applications, as in the bioremediation of petroleum polluted environments.
Figure 1:Pseudomonas aeruginosa strains isolated from Romanian environments polluted with petroleum hydrocarbons.
Acknowledgement
The study was funded by project no. RO1567-IBB05/2019 from the Institute of Biology Bucharest of Romanian Academy.
References
10. Ali CH, Qureshi AS, Mbadinga SM, Liu JF, Yang SZ, et al. (2016) Organic solvent tolerant lipase from Pseudomonas aeruginosa FW_SH-1: purification and characterization. JSM Enzym Prot Sci 1: 1005.
17. Isken S, de Bont JAM (1998) Bacteria tolerant to organic solvents. Extremophiles 2(3): 229-238.
19. Matilla MA (2018) Problems of solventogenicity, solvent tolerance: an introduction. In: Krell T (Ed.), Cellular Ecophysiology of Microbe: Hydrocarbon and Lipid Interactions, Handbook of Hydrocarbon and Lipid Microbiology, Springer, Cham, Germany, pp. 327-334.
21. Varjani SJ (2017) Microbial degradation of petroleum hydrocarbons. Bioresour Technol 223: 277-286.
23. Stancu MM (2015) Isolation and characterization of new marine oil-degrading bacteria. Rom Biotechnol Lett 20: 10316-10326.
© 2019 Stancu Mihaela Marilena. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






