- Submissions

Full Text
Journal of Biotechnology & Bioresearch
Enhanced Succinic Acid Production in Escherichia coli by Model-Guided Metabolic Gene Knockout of pflA Using Glucose Carbon Source
Bashir Sajo Mienda1*,Saidu Jibril2, Umar Ali3, Aminu Muhammad4, Rabiu Salihu1 and Aliyu Adamu5
1 Department of Microbiology & Biotechnology, Federal University Dutse, Nigeria
2 Department of Chemical Sciences, Federal University Kashere, Nigeria
3 Department of Chemistry, SuleLamido University Kafin Hausa, Nigeria
4 Department of Pure and Industrial Chemistry, Bayero University Kano, Nigeria
5 Department of Microbiology, Kaduna State University, Nigeria
*Corresponding author:Bashir SajoMienda, Department of Microbiology & Biotechnology, Faculty of Science, Federal University Dutse, PMB 7156 Ibrahim Aliyu Bypass, Dutse, Jigawa State, Nigeria
Submission: May 30, 2018;Published: July 09, 2018

Volume1 Issue1July 2018
Abstract
Succinic acid is an important platform and/or commodity or specialty chemical with a broad range of applications. The metabolic role of pyruvate formatelyase A (pflA) in relation to succinate production in Escherichia coli under anaerobic conditions from glucose substrate remained largely unspecified. Herein we identified pflAgene for the first time, as a novel gene knockout target for increasing succinate production in E. coli. Guided by E. coli reconstruction iJO1366, we engineered the E. coli host metabolism by deleting the pflA, thereby causing the up-regulation of glyceraldehyde- 3-phosphate dehydrogenase (GAPDH), which hypothetically increases the generation of NADH and the pool of phosphoenolpyruvate (PEP) in the central carbon metabolism, required for succinate production. This strategy produced succinic acid that is 4.78 fold (0.28g l-1 in 1day) from glucose substrate. This work elucidatesfor the first time that pflA is a novel gene deletion target for increasing succinic acid production in E. coli under anaerobic conditions. In addition, these results highlight the power of metabolic model in identifying novel gene deletion target and ultimately driving novel biological discovery.
Keywords: Escherichia coli genome-scale model;Metabolic gene knockout prediction;Pyruvate formatelyase (pflA); Glucose carbon source; Enhanced succinate production
Introduction
Microbial fermentation for succinic acid production has been pursued in recent time because it is considered cheaper and environmentally friendly approach than its petroleum based chemical production from maleic anhydride [1]. Recently, several bacteria, such as Anaero biospirillum succiniciproducens, Actinobacillus succinogenes and Mannheimia succiniciproducens have been established to produce succinic acid as a major fermentation product [2]. However, these strains require complex organic nutrients that increase the costs of productions, purifications and the waste disposal which ultimately add to process costs and complexity [2,3]. Escherichia coli have been known to naturally carry out mixed acid fermentation with succinic acid as a minor fermentation product among others. As a specialty and/or commodity chemical, succinic acid has invaluable applications, such as a precursor for various chemicals, including green solvents and biodegradable plastics, it can also be used as an iron chelator and a supplement to many foods and pharmaceuticals [1,2]. Succinic acid has also been listed by the U.S. Department of Energy (DOE) among the 12 top biobased building block chemicals that can be produced by microbial fermentation [4,5]. Several numbers of metabolically engineered E. coli strains were constructed, with or without foreign genes for enhanced succinate production using glucose substrate [2]. While others, used mineral salt medium to produce succinate in metabolically engineered E. Colistrains by knocking out pyruvate formatelyase B (pflB) [3]. pflB was previously designated as a formate acetyltransferase I, which its deletion under anaerobic conditions blocks formate formation and increase succinate production [3,6]. The disruption of pyruvate formatelyase (pfl) was established to increase D-lactate production in E. coli under micro-aerobic conditions [7]. The deletion of pflA for increasing succinic acid production has not yet been elucidated.
The proliferation of E. coli genome scale models (GEM) [8,9] have facilitated the application of systems metabolic engineering to increase the production of desired compounds. One of the application of E. coli GEM is in metabolic engineering interventions and targeted biological discovery among others [10]. Although recently a number of studies have shown that GEMs of E. coli can be deployed for metabolic gene knockouts in increasing succinic acid production, only few studies reported the use of E. coli GEM to guide metabolic engineering [11]. The use of E. coli genome scale metabolic model to guide future experimental studies would offer considerable help in reducing the time and costs of a targeted biological discovery. Direct experimental trial and error approach was employed to increase D-lactate production in E. coli, following the metabolic gene knockout of the entire pyruvate formatelyases (pflA, pflB, pflC and pflD) [7]. These deletions, particularly of pflA and pflB were established to cause up the regulations of glyceraldehyde- 3-phosphate dehydrogenase (GAPDH) andpyruvate kinase (PYK) glycolytic enzymes, thereby generating NADH that facilitated increase purely D-lactate production in E. coli [7]. But the specific role of pflA when deleted for increasing succinic acid production has not been reported. The current study hypothesizes that pyruvateformatelyase activating enzyme1 (encoded by pflA) could be a novel gene knockout target for increasing succinate production in E. coli. To facilitate and expedite our efforts, we leveraged predictive computational modeling of metabolism and model-guided analysis of experimental data. I applied constraint-based metabolic modeling by deleting the pflAgene using both the substrates to increase succinate production in E. coli. This is because researchers have reported the successful application of metabolic models of E. coli to engineer strains that produce succinic acid [1,11]. Herein we report for the first time the model-guided identification of pflA as a novel gene deletion target for increasing succinic acid production in E. coli as initially hypothesized. An accurate E. coli GEM [9] and Minimization of Metabolic Adjustment (MOMA) algorithm [12] in the OptFlux software platform [13] were utilized for the prediction of the target, and subsequently confirmed experimentally. It is worth mentioning that the current study achieved construction of an E. coli mutant strain designated as BSM3 with succinic acid production titer that is 4.7 fold (0.28g l-1 in 1 day) from glucose substrate. This study informs other studies that pflA is a novel target that can be deleted to increase succinic acid production in E. coli and probably beyond.
Materials and Methods
In silico analysis of gene knockout
Escherichia coli genome scale stoichiometric model iJO1366 [9] was employed for the in silico simulation of gene deletion by using Minimization of Metabolic Adjustment (MOMA) algorithm [12] with OptFlux software platform [13]. The E. coli iJO1366 model has been tested and proven to be predictive for computations of growth rates and metabolite excretion rates from a range of substrates and genetic conditions [9,14]. MOMA was described as a flux based analysis technique that employs quadratic programming to search for the nearest point in the feasible solution space of the mutant model in relations to its wild-type optimal point feasible solution space [12]. The OptFlux software platform is an in silico metabolic engineering (ME) platform that was implemented using the Java programming, which contains MOMA as a simulation algorithm. Flux balance analysis (FBA) was used for all phenotype simulations. All the simulation of the mutant and the wild-type models were performed using the OptFlux software version 3.07 Glucose was used as solitary carbon source under anaerobic conditions. The substrate uptake rate was constrained to a maximum of 18.5mmolgDW- 1 h-1 whereas the corresponding oxygen uptake rate was set to zero, as the environmental conditions are anaerobic. These values were selected based on closely established experimental observations on aerobic and anaerobic growth in E. coli [15,16].
Bacteria and plasmid
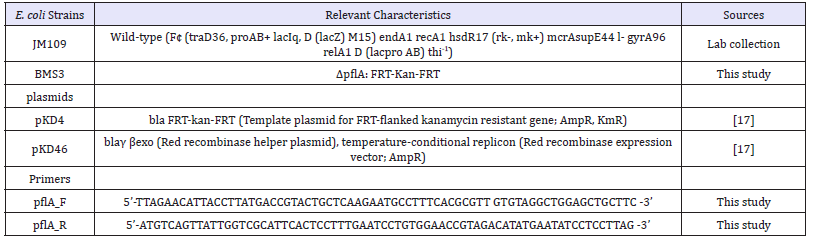
E. coli JM109 (F¢ (traD36, proAB+ lacIq, D (lacZ) M15) endA1 recA1 hsdR17 (rk-, mk+) mcrAsupE44 l- gyrA96 relA1 D (lacproAB) thi-1) was used for maintenance of the pkD4 and pkD46 plasmids. The plasmids were used strictly following the method described previously [17]. The plasmid pKD4 was extracted from E. coli JM109 using the QIAprepMiniprep kit according to the manufacturer’s specifications.
Media chemicals and other reagents
E. coli cells used in this study were grown in LB medium containing 0.5% yeast extract (Difco), 0.5%NaCl and 1% Bactotryptone (Difco) without or with antibiotics at the concentrations of 100μg/ ml ampicillin and 30μg/ml of Kanamycin. L-arabinose, and glucose were obtained from Sigma Aldrich. KAPA HiFiHotstart Ready Mix (2X) was from KAPA BIOSYSTEMS. Agarose was purchased from (Sigma Aldrich).
PCR primers
The E. coli pflA gene sequence was used to design forward and reverse primers with pKD4 template plasmid sequence. The primers had 50-nt 5’ extension including the gene initiation codon (H1) and 20-nt sequence (p1) as described previously [17,18]. Table 1 gives the details of the primers used in this study.
Generation of PCR fragments
PCR reactions were carried out in an Eppendorf thermo cycle using 25μl reactions containing 12.5μl of KAPA HiFiHotstart Ready Mix (2X), 1μl of pKD4 template DNA, 1.0μl of each primer. Reactions were performed for 30 cycles: 95 °C for 3min, 98 °C for 20 secs, 55 °C for 15 sec, 72 °C for 1:30 sec, 72 °C for 60 sec and cooling at 4 °C . PCR products were purified using SV gel and PCR clean up system (Promega, USA), according to the manufacturer’s protocol. Then, the PCR products obtained were analyzed by 1% agarose gel-electrophoresis using 1X Tris-acetate buffer.
Electroporation and mutant selection
E. coli JM109 harboring the λ-Red helper plasmid pKD46 was grown in 100ml of LB medium with ampicillin and 1mM L-Arab inose at 30 °C to an OD600 of 0.3. Competent cells for electroporation were prepared as described previously [19]. A 1.0μl (400ng) aliquot of the PCR fragment was mixed with 50μl of competent cell in an ice-cold Eppendorf electroporation cuvette (0.2cm). Electroporation was performed at 2.5KV with 2mF and 600Ω and was followed by immediate addition of 1ml of SOC medium (0.5% yeast extract (Difco), 2% Bactotryptone (Difco), 2.5mMKCl, 10mM NaCl, 10mM MgCl2, 10mM MgSO4 and 20mM glucose) with 1mM L-arabinose. The SOC medium mixed with the electroporated cells was incubated for 2 hours at 37 °C. Selection of kanR transformant was followed immediately by spreading one-tenth portion of the electroporated cells onto kanamycin agar plates as described by Baba and colleagues [18]. To test for accurate mutational inactivation or correct chromosomal structure, 20μl PCR verification method was conducted with kanamycin specific primers K1 and K2 as described earlier [17].
Table 1:Escherichia coli K12 strains, plasmids and primers used in this study.
Anaerobic fermentation
Bacterial cells starter culture was made by growing the cells in 10ml LB medium with shaking at 200rpm at a temperature of 37 °C. One milliliter of seed culture was used to inoculate a 125ml butyl rubber stoppered serum vial, which contained 100ml of fermentation media as described by Lee and colleagues [1]. The fermentation media used contained the following ingredients (per liter): yeast extract=5g; glucose=9g (50mM); NaHCO3=10g; NaH2PO4. H2O=8.5g; K2HPO4=15.5g (pH=7.0). Anaerobic conditioning was established by filling the headspace with N2 and addition of Na2S.9H20 (final 1mM). Cells were cultivated under anaerobic conditions at 37 °C with shaking at 200rpm for 1 to 3 days unless otherwise stated.
Analytical procedure
The concentrations of glucose, ethanol and organic acids (lactate, formate, and succinate) were quantified by high performance liquid chromatography using the Agilent 1260 Infinity (Agilent Technologies, USA). The HPLC Agilent, equipped with an RI detector and a 300×7.88mm Aminex HPX-87H ion-exchange column (Bio-Rad laboratories, USA), was used for these purposes. The culture supernatant was passed through a syringe filter (pore size of 0.2μm) after centrifugation at 10,000×g for 10min and stored at -20 °C for analyses. For operating conditions to optimize peak separation for D-glucose substrate, the column was eluted isocratically at 47 °C with a flow rate of 0.6ml min-1 using 0.01NH2SO4 as the mobile phase, following the description in their original documentations [1]. To quantify cell growth, the optical density of the cell cultures was measured at 600nm using a GENESYS 105 VIS spectrophotometer (Thermo scientific, USA).
Results and Discussion
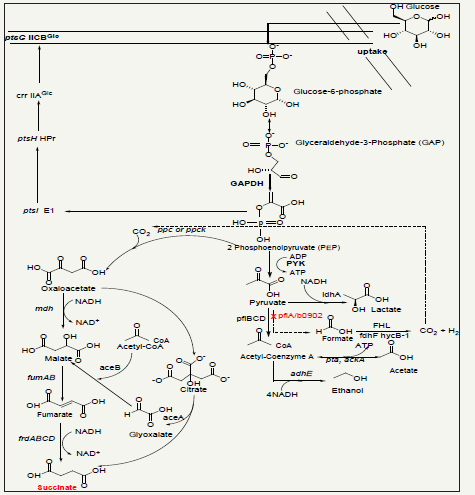
Escherichia coli genome-scale metabolic model could help in identifying novel gene deletiontargets for increasing succinic acid production. In this study, we initially hypothesized that the deletion of pflA gene in E. coli could increase succinic acid production under anaerobic condition using glucose as substrate. The predicted results obtained with the E. coli GEM using glucose substrate shows decrease in succinate production (95% of the wild-type model) following the deletion of pflA (Table2), but the experimental validation with the same substrate proves otherwise, which is nearly 4.7 fold in 1 day (0.28gl-1) and 3.2 fold in 3 days (0.30g l-1) and their corresponding parent strains produced only 0.058g l-1 and 0.096g l-1 respectively (Table 3). On one hand, the hypothesis that the pflAgene deletion could increase succinic acid production in E. coli under anaerobic condition have been experimentally validated while on the other hand inconsistencies exists in model’s predictions results relative to the experimental outcomes.It was reported previously [7] that pyruvate is mainly cleaved via pyruvate formatelyase (pfl) to form formate and acetyl-coA [19]. The specific deletion of pflA is not clearly specified in relation to succinate production in E. coli under anaerobic conditions, but it was established to cause up regulation of GAPDH and PYK when compared to their parent strains [7]. On the basis of these findings, the plausible hypothetical mechanism for the increased succinate production in strain BMS3 (ΔpflA) could be theoretically attributed to the up regulation of GAPDH and PYK (Figure 1). The up regulation of PYK was previously described as a gluconeogenic process using NADH-linked malic enzyme that increases succinate production in E. coli [2,20].
Table 2:In silico prediction results.
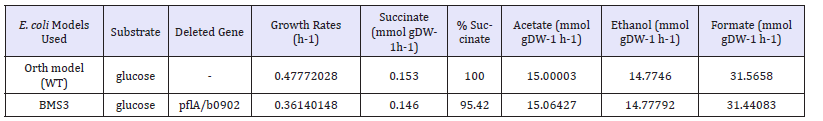
Figure 1:Metabolic pathways involved in the anaerobic utilization of glucose in E. Coli [2,3] and metabolic pyruvate formatelyase (pflA) gene knockout strategy employed for producing succinate in this study.
The anaerobic conditions established during the fermentative production of succinate in BMS3 strain, might have led to stepping up of glycolysis, and ATP may have been generated via substrate level phosphorylation. This phenomenon coupled with pflAgene knockout might have theoretically lead to the generation of additional NADH through the up regulation of GAPDH which resulted in excess NADH/NAD+ ratio in BSM3 mutant strain. Correspondingly, the succinate production increase was achieved to meet the requirements of redox balance and the energy production through glycolysis. The production of succinate in E. coli using glucose under anaerobic condition was previously established to consume 2 molecules of NADH per succinate produced [21]. The enzymes in bold (GAPDH and PYK) were up regulated following the pflA gene, which could have been responsible for increased succinate production. Relevant genes and enzymes involved in succinate productions are shown in italics. Broken lineindicate additional CO2 generated following the oxidation of formate to CO2 and H2 by formate hydrogen lyase (FHL). The additional CO2 generated may have contributed to addional CO2 fixation by ppcfor the PEP conversation to OAA, and step wisely converted to succinate. Abbreviations: GAPDH, glyceraldehyde- 3-phosphate dehydrogenase; PYK, pyruvate kinase; PEP, phosphenol pyruvate; ppc, phosphoenolpyruvate carboxylase; mdh, malate dehydrogenase; fumABC, fumarate hydratases; frdABCD, fumarate reductases
The single deletion of the pflA gene might have increases the NADH pool, because lactate dehydrogenase (ldhA) and alcohol dehydrogenase (adhE) genes were not deleted in our mutant strain BMS3, to primarily show that the succinate production increase is caused by pflA gene knockout. Correspondingly, fermentative profile of the mutant strain BMS3 indicated a clear increase in succinate, lactate and ethanol production from the wild-type using glucose substrate (Table 3).
Table 3:Fermentative succinate production profile from glucose by engineered E. coli strain BSM3 (ΔpflA) with its wildtype strain used in this studya
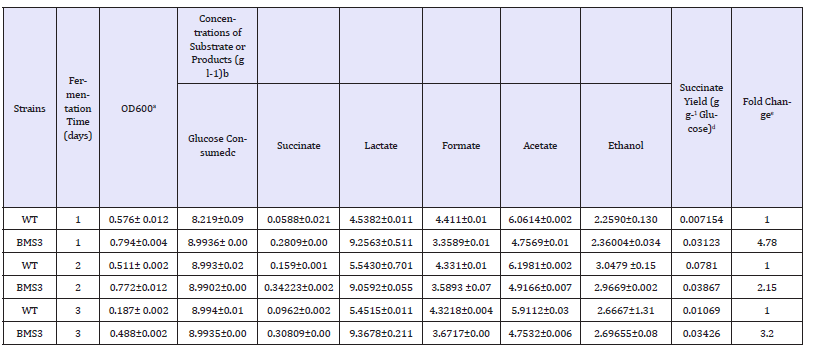
a Data represent the averages of three samples (mean ± standard deviations) taken from days of anaerobic fermentation cultures supplemented with 9 g l-1 of glucose unless otherwise specified.
b Anaerobic vial fermentation on 9g l-1 initial glucose for 1 to 3 days
c Calculated by subtracting the initial glucose concentration from the residual glucose concentration
d Calculated as (g l-1 of succinate produced) / (g l-1 of glucose consumed)
e Calculated as succinate titer in mutant / succinate titer in the wild-type.
The increase in ethanol and lactate production in this study would not have been possible in the cell without increase in NADH pool, because production of lactate and ethanol are established NADH linked phenomenon [22-24]. Therefore, the additional NADH generated has been used in increasing succinate production in the mutant strain BMS3. This study clearly establishes that single pflA gene knockout is solely responsible for the increased in succinate production under anaerobic conditions in E. coli from glucose substrate.Another interesting reason that could be possible for increasing succinate production in the mutant strain BMS3 with the glucose substrate is the activities of phosphoenolpyruvate carboxylase( PPC) and acetate kinase (ACK). These two enzymes were previously reported to have increased activities following the deletion of pflA [7]. The PPC is the first enzyme for succinate production in E. coli, therefore, based on this enzyme activity, significant succinate production is achieved in E. coli using glucose under anaerobic conditions. In a similar study reported for purely D-lactate production under micro-aerobic conditions, pflAgene knockout in E. coli did not increases succinate production because of CO2and PEP shortage, as the condition for D-lactate production in E. coli was micro- aerobic (limited amount of O2 enters the system) [7].
In contrast, the mutant strain BSM3 achieved increase in succinate production with the same pflA gene knockout, because the fermentative condition employed in this study is completely anaerobic, supplying additional CO2 for PPC and increasing the pool of PEP by the up-regulation of GAPDH (Figure 1).The blocking of the entire pyruvate assimilation pathway under micro-aerobic and anaerobic conditions by inactivating the pflABCD, could cause the shortage of acetyl-CoA (AcCoA) in E. coli [7]. Although, under fermentative condition, pflB is the predominant route for pyruvate conversion to acetyl-CoA synthesis[25]. pflB was established to be responsible for formate formation, which can be subsequently cleaved to CO2and H2 by formate hydrogen lyase (fdhF/hycB-1) [26] (Figure 1). The reason why we decided not to delete pflB in this study is because we need formate generation which could be subsequently converted to CO2and H2, and additional CO2 is required for the efficient functioning of PPC to convert PEP to OAA, which could ultimately be used for succinate production.Reduced formation of formate and acetate on glucose substrate was demonstrated by our mutant strain BMS3 (Table 3), because of the deletion of pflactivating enzyme (pflA). This is because pflB in our mutant strain might have contributed to tformation of formate, and fdhF/hycB-1 (Figure 1) could have converted the formate to CO2. This could be an additional reason why reduced formate formation was observed when glucose is used as substrate (Table 3). The acetate kinase (ACK) and phosphotransacetylase (PTA) designated as ACK-PTA pathway in E. coli is related to AcCoA pool [7]. This pathway was established to have two directions: one direction is to produce 1mol of ATP by acetate excretion, while the other direction is by consuming 1mol of ATP by utilizing acetate to produce intracellular AcCoA [7]. Our mutant strain BMS3 is deficient in pflA, therefore AcCoA formation via pflA will be minimized, ACK-PTA reactions could occur in the direction favoring AcCoA formation and utilizing acetate and ATP for biomass formation and energy maintenance. This could be the plausible reason why even after the deletion of the pflAgene, yet acetate and formate were produced as fermentation end product in our mutant strain BMS3 (Table 3). Taken together, the model-guided deletion of the pflA gene in E. coli for succinate production from glucose substrate described in this study, is a step forward towards understanding the metabolic role of this deletion and suggests that model can drive novel biological discovery.
The inconsistencies in the results for succinate predicted flux and observed experimental measurements reported in this study represent true biological gaps (incomplete knowledge gaps) in the reconstruction iJO1366) [27]. In addition, models contains missing regulatory processes, and thus could open up further opportunities for novel biological discovery [28-30] on the missing pflA gene function in relation to E. coli metabolism and succinate production under anaerobic conditions. In addition, the current study also clearly established that novel gene deletion targets could be identifiedby combining expert knowledge, model-guided and/or systems based metabolic engineering strategies for microbial strain improvement. This strategy could also offer a considerable biological insight for strain improvement for the production of value-added compounds, such as succinate, xylitol, ethanol etc., from renewable feedstock such as glucose.
Conclusion
The current study hypothesizes that the deletion of pflA in Escherichia coli could increase succinic acid production using glucose carbon source. This hypothesis was predicted using E. coli GEM and later experimentally confirmed to have increased succinic acid production from glucose carbon source, suggesting that pflA could be considered as a novel gene deletion target that could increases succinic acid production in E. coli, and could ultimately guide future metabolic engineering strategies for increasing the acid production and/or any other chemical that requires additional NADH for its production in E. coli and beyond.
References
- Lee SJ, Lee DY, Kim TY, Kim BH, Lee J, et al. (2005) Metabolic engineering of Escherichia coli for enhanced production of succinic acid, based on genome comparison and in silico gene knockout simulation. Appl Environ Microbiol 71(12): 7880-7887.
- Zhang X, Jantama K, Moore JC, Jarboe LR, Shanmugam KT, et al. (2009) Metabolic evolution of energy-conserving pathways for succinate production in Escherichia coli. Proc Natl Acad Sci U S A 106(48): 20180- 20185.
- Zhang X, Jantama K, Shanmugam KT, Ingram LO (2009) Reengineering Escherichia coli for succinate production in mineral salts medium. Appl Environ Microbiol 75(24): 7807-7813.
- Choi S, Song CW, Shin JH, Lee SY (2015) Biorefineries for the production of top building block chemicals and their derivatives. Metab Eng 28: 223-239.
- Thakker C, Martínez I, San KY, Bennett GN (2012) Succinate production in Escherichia coli. Biotechnol J 7(2): 213-224.
- Zhang X, Shanmugam KT, Ingram LO (2010) Fermentation of glycerol to succinate by metabolically engineered strains of Escherichia coli. Appl Environ Microbiol 76(8): 2397-2401.
- Zhu J, Shimizu K (2004) The effect of pfl gene knockout on the metabolism for optically pure D-lactate production by Escherichia coli. Appl Microbiol Biotechnol 64(3): 367-375.
- Feist AM, Henry CS, Reed JL, Krummenacker M, Joyce AR, et al. (2007) A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information. Mol Syst Biol 3: 121.
- Orth JD, Conrad TM, Na J, Lerman JA, Nam H, et al. (2011) A comprehensive genome-scale reconstruction of Escherichia coli metabolism--2011. Mol Syst Biol 7: 535.
- McCloskey D, Palsson BO, Feist AM (2013) Basic and applied uses of genome- scale metabolic network reconstructions of Escherichia coli. Mol Syst Biol 9: 661.
- Wang Q, Chen X, Yang Y, Zhao X (2006) Genome-scale in silico aided metabolic analysis and flux comparisons of Escherichia coli to improve succinate production. Appl Microbiol Biotechnol 73(4): 887-894.
- Segre D, Vitkup D, Church GM (2002) Analysis of optimality in natural and perturbed metabolic networks. Proc Natl Acad Sci U S A 99(23): 15112-15117.
- Rocha I, Maia P, Evangelista P, Vilaça P, Soares S, et al. (2010) OptFlux: an open-source software platform for in silico metabolic engineering. BMC Syst Biol 4: 45.
- Feist AM, Zielinski DC, Orth JD, Schellenberger J, Herrgard MJ, et al. (2010) Model-driven evaluation of the production potential for growth-coupled products of Escherichia coli. Metab Eng 12(3): 173-186.
- Varma A, Boesch BW, Palsson BO (1993) Stoichiometric interpretation of Escherichia coli glucose catabolism under various oxygenation rates. Appl Environ Microbiol 59(8): 2465-2473.
- Varma A, Palsson BO (1994) Stoichiometric Flux Balance Models Quantitatively Predict Growth and Metabolic By-Product Secretion in Wild- Type Escherichia coli W3110. App Environ Microbiol 60(10): 3724-3731.
- Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97(12): 6640-6645.
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, et al. (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2: 2006.0008.
- Sharan SK, Thomason LC, Kuznetsov SG, Court DL (2009) Recombineering: a homologous recombination-based method of genetic engineering. Nat Protoc 4(2): 206-223.
- Mienda BS, Shamsir MS (2015) In silico deletion of PtsG gene in Escherichia coli genome-scale model predicts increased succinate production from glycerol. J Biomol Struct Dyn 33(11): 2380-2389.
- Cheng KK, Wang GY, Zeng J, Zhang JA (2013) Improved succinate production by metabolic engineering. Biomed Res Int 2013: 538790.
- Dharmad Y, Murarka A, Gonzalez R (2006) Anaerobic fermentation of glycerol by Escherichia coli: a new platform for metabolic engineering. Biotechnol Bioeng 94(5): 821-829.
- Mattam AJ, Clomburg JM, Gonzalez R, Yazdani SS (2013) Fermentation of glycerol and production of valuable chemical and biofuel molecules. Biotechnol Lett 35(6): 831-842.
- Clomburg JM, Gonzalez R (2013) Anaerobic fermentation of glycerol: a platform for renewable fuels and chemicals. Trends Biotechnol 31(1): 20-28.
- Durnin G, Clomburg J, Yeates Z, Alvarez PJ, Zygourakis K, et al. (2009)Understanding and harnessing the microaerobic metabolism of glycerol in Escherichia coli. Biotechnol Bioeng 103(1): 148-161.
- Blankschien MD, Clomburg JM, Gonzalez R (2010) Metabolic engineering of Escherichia coli for the production of succinate from glycerol. Metab Eng 12(5): 409-419.
- Orth JD, Palsson BO (2010) Systematizing the generation of missing metabolic knowledge. Biotechnol Bioeng 107(3): 403-412.
- Guzman GI, Utrilla J, Nurk S, Brunk E, Monk JM, et al. (2015) Model-driven discovery of underground metabolic functions in Escherichia coli. Proc Natl Acad Sci U S A 112(3): 929-934.
- Monk J, Palsson BO (2014) Genetics. Predicting microbial growth. Science 344(6191): 1448-1449.
- Brien OEJ, Monk JM, Palsson BO (2015) Using genome-scale models to predict biological capabilities. Cell 161(5): 971-987.S
© 2018 Bashir Sajo Mienda. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






