- Submissions
Full Text
Interventions in Obesity & Diabetes
Alterations of Mitochondrial Functions and DNA in Diabetic Cardiomyopathy of CCK1 Receptors-Deficient Rats
Abdelbary Prince1,2, Magdy A Ghoneim13, Abdallah M El-Ebidi4, Hala A Mousa5 and Jin Han2*
1Department of Biochemistry, Cairo University, Egypt
2Department of Physiology, College of Medicine, Inje University Korea
3Department of Biochemistry, King Abdulaziz University, Saudi Arabia
4Department of Medical Biochemistry and Molecular Biology, Aswan University, Egypt
5Department of Internal Medicine and Nephrology, Aswan University, Egypt
*Corresponding author: Jin Han, National Research Laboratory for Mitochondrial Signaling, Department of Physiology, College of Medicine, Cardiovascular and Metabolic Disease Center, Inje University 633-165 Gaegeum-Dong, Busanjin-Gu, Busan 613-735, Korea
Submission: February 09, 2018; Published: February 22, 2018

ISSN: 2578-0263Volume1 Issue2
Abstract
Many data has accumulated supporting the involvement of oxidative stress in the development of diabetic cardiomyopathy. The current study was carried out to investigate the effect of diabetic oxidative stress on cardiac mitochondrial functions to figure out potential biomarkers and therapeutic targets for early diagnosis and treatment of diabetic cardiomyopathy. For this purpose, a total number of 18 male Otsuka Long-Evans Tokushima Fatty (OLETF) rats as diabetic group and age-matched Long-Evans Tokushima Otsuka (LETO) as control group were used in the present study.
Rats (both LETO and OLETF) were anesthetized and thoracotomy was performed, and hearts were per-fused. Furthermore, for mitochondrial isolation 12 hearts were used (6 for flow cytometry and another 6 for measurement of oxygen consumption). Additionally, to carry out DNA fragmentation assay and 8-OHdG measurement, 6 hearts were used for mitochondrial DNA extraction.
The obtained findings can be summarized in that electron microscopy of mitochondria from diabetic OLETF hearts revealed increases in both size and number Moreover, mitochondrial DNA fragmentation percentage and oxidized mitochondrial DNA were higher in diabetic hearts when compared with control group, and demonstrated impaired mitochondrial function in diabetic group. In addition, data showed decrease in ETC respiration as measured by the alteration in respiratory control ratio (RCR) of diabetic OLETF heart mitochondria. It can be concluded that mitochondrial ROS are involved in the progression of diabetic cardiomyopathy. This can be explained in the light of the progression of mitochondrial dysfunction and increasing level of oxidative stress during diabetes in cardiac tissue.
Keywords: Mitochondria, Diabetes mellitus, Cardiac myocytes, obesity, oxidative stres
Introduction
Diabetes mellitus is a complicated, chronic disorder characterized by either insufficient insulin production by pancreatic β-cells or by cellular resistance to insulin. Oxidative stress has been suggested as a contributory factor in the pathogenesis of diabetes [1].
Type 2 diabetes mellitus (T2DM) is a metabolic disease characterized by elevation of blood glucose concentration, lipid abnormalities and vascular complications. Insulin resistance and pancreatic β-cell insufficiency with respect to insulin production are major features in the progression of T2DM [2,3]. Obesity is defined as excess body fat, determined by a surrogate measure called body mass index (BMI), defined as weight in kilograms over height in meters squared. Abdominal obesity due to excess visceral fat is associated with an increased risk of developing cardiovascular disease [4]. Moreover, excess visceral fat is linked to an increased risk of metabolic syndrome, which includes a greater risk of developing type 2 diabetes mellitus (NHLB, 2006) associated with cardio-metabolic disorders [5].
Today, over 95% of all diabetes is type 2 and it occurs in children and adolescents as well as adults. In an individual with a BMI of 30, the risk of diabetes type 2 is increased 60- to 80-fold in comparison to lean individuals. Meanwhile, the risk for heart disease is only 4-to 6-fold increased at a BMI of 30. The association of diabetes type 2 with obesity goes beyond typical risk factors and justifies naming the disease diabesity. However, insulin treatment failed to reduce cardiovascular mortality [6]. Several lines of evidence implicate mitochondrial defects as a major factor in diabetes mellitus [7]. Diabetes mellitus has been proposed to result from defects in the glucose sensor for insulin secretion [8], insulin resistance [9], and from defective modulation of the β-cell KATP channels [10]. All these three factors can be tied together through mitochondrial energy production.
Furthermore, type II diabetes patients consistently show a down-regulation in the expression of nDNA encoded mitochondrial genes, in association with alterations in the levels ofthe peroxisome- proliferation-activated receptor-y (PPARy)-coactivator 1 (PGC- 1) [11], a major regulator of mitochondrial biogenesis and fat oxidation [12].
Diabetes mellitus is also seen in Friedreich ataxia as the result of inactivation of the frataxin gene. Frataxin binds iron in the mitochondrial matrix, thus minimizing mitochondrial oOH production. The loss of the frataxin protein results in excessive ROS generation which inactivates all mitochondrial iron-sulfur- containing enzymes. Thus increased mitochondrial ROS production and decreased mitochondrial OXPHOS must be the cause of diabetes in Friedreich ataxia [13]. The importance of mitochondrial defects in β-cell insulin secretion deficiency has been confirmed in two mouse models. In the first, the mitochondrial transcription factor T-fam was inactivated in the pancreatic β-cells. This resulted in increased blood glucose and the progressive decline in β-cell mass by apoptosis [14]. In the second, the ATP-dependent K+- channel (KATP) affinity for ATP was reduced, resulting in a severe reduction in serum insulin, severe hyperglycemia with hypoinsulinemia, and elevated 3-hydroxybutyrate levels [10]. These models demonstrate that mitochondrial ATP production is critical in the signaling system of the β-cell to permit insulin release [15].
The aim of this study is to investigate the cardiac mitochondrial functions in relation to diabetic oxidative stress of knockout obese rats. This may help to discover novel biomarkers and to identify new therapeutic targets for diabetic cardiomyopathy.
Materials and Methods
Experimental animals
Nine male Otsuka Long-Evans Tokushima Fatty (OLETF) rats of 8 weeks age and nine age-matched Long-Evans Tokushima Otsuka (LETO) rats were obtained from the Animal Center of Tokushima Research Institute (Otsuka Pharmaceutical, Tokushima, Japan) and maintained until they reached an appropriate age for the experiment. OLETF rats lacking CCK1 receptors are hyperphagic following weaning and become obese. OLETF rats have impaired glucose tolerance by 5 weeks of age after that the degree of glucose intolerance increases and became clearly hyperglycemic and hyperinsulinemic (i.e. develop type 2 diabetes). All rats had free access to standard laboratory chow and tap water, and were taken care of under the specifications outlined in the Guiding Principles for the Care and Use of Laboratory Animals—Approved by the Institutional Animal Care and Use Committee of Inje University.
All investigations were performed according to the "NIH Guide for the Care and Use of Laboratory Animals" (National Institutes of Health Publication No. 85-23, Revised 1996) and approved by the Institutional Animal Care and Use Committee of Inje University College of Medicine, Korea.
Experimental design
Thirty-five weeks old male OLETF rats (696±52g) used as diabetic group and 35 weeks old male LETO rats (480±26g) used as control. Three rats from each group were used for mitochondrial DNA extraction and 6 rats from each group were used for mitochondrial isolation.
Sampling: At 35 weeks of age all animals were sacrificed under sodium pentobarbital (80mg/kg, i.p.) anesthesia and heparin sodium (500IU/kg. i.p.) as anticoagulant and heart and thoracic aorta were dissected out.
Electron microscopy: The morphology of the mitochondria was assessed using an electron microscope (JEOL 1200 EX2, Japan). The procedures were done according to the method of Rajapakse et al, [16]. In brief, samples were fixed in 2.5% glutaraldehyde then washed three times for 10 min each with 0.1M Millonig phosphate buffer, followed by post-fixation for 1 h in 2% (w/v) osmium tetroxide. Samples were then washed 3 times for 10 minutes each with buffer followed by dehydration in a graded series of ethanol solutions (25%, 50%, 85%, 95%, and 100%) for 10min each. Samples were further dehydrated twice for 10 minutes each in propylene oxide and infiltrated for 2h in a 1:1 mixture of propylene oxide and purr-resin, then overnight in a 1:2 mixture of propylene oxide and purr-resin. Finally, the samples were infiltrated in pure purr-resin for 6h before embedding and curing at 70 0C overnight. Electron microscopy was performed using a JEOL 1200 EX2, Japan transmission electron microscope.
Flow cytometry of mitochondrial superoxide: Double staining of mitochondria was performed according to Spallarossa et al. [17]. For evaluation of mitochondrial ROS production, we loaded the isolated heart mitochondria with the fluorescent probes H2DCF- DA and Mito-Tracker Red CM-H2XRos, respectively. Briefly, six isolated rat hearts from each group (both LETO and OLETF) were manually homogenized, using medium fitting glass-teflon Potter- Elvehjem homogenizer in mitochondrial isolation buffer (MIB). The resultant homogenate was centrifuged at 1500xg for 5minutes at 4 0c. The supernatant was centrifuged again at 10,000xg for 10minutes and the mitochondria-enriched pellet was resuspended in mitochondrial isolation buffer. Mitochondria were labeled by the addition of Mito-Tracker Red CMXRos to the suspended mitochondrial pellet (100nM final concentration; Molecular Probes, Eugene, OR) and H2DCF-DA was added to final concentration of 5|iM according to manufacturer's recommendation. The mixture was incubated for 10min in the dark at room temperature, and then fluorescence was measured by flow cytometry analysis with a FACS-SCAN apparatus (FACSCalibur; BD Biosciences, USA).
Mitochondrial DNA fragmentation assay: The DNA fragmentation assay was performed according to the method of Herrmann et al. [18]. Mitochondrial DNA isolation was performed as described by Rudolf et al. [19] using mtDNA Extractor CT Kit. In brief, 50mg of minced cardiac tissue from six isolated hearts from each group (both LETO and OLETF) were homogenized with scissors in 1mL of the homogenizing buffer with a glass homogenizer by up- and down of the pestle on ice until the pestle reaches the bottom of the glass well. The homogenate was centrifuged at 1000xg for I minute at 4 0C. The supernatant was recentrifuged at 10000xg for 10 minutes at 4 oc. DNA extraction solution I was added to the precipitate and kept on ice for 5 minutes. DNA extraction solution II was added and kept on ice for 5 minutes. Cold DNA extraction solution III was added then kept on ice for 5 minutes. Centrifugation was performed at 12000xg for 5 minutes at 4 oc. Supernatant was transferred to new microtube then sodium iodide solution was added. Isopropanol was added then centrifugation was performed at 12000xg for 10 minutes at room temperature. The precipitate was washed using washing solution then centrifugation was applied at 12000 xg for 5 minutes at room temperature and this step was repeated twice. The pellet was dried under vacuum and then re-suspended in TE buffer (pH 8.0). The mitochondrial DNA quantity and purity were assessed spectrophotometrically using NanoDrop-1000 spectrophotometer and ND-1000 software. Agarose gel electrophoresis (1.5%) of mtDNA was performed as described by Sambrook & Russel [20].
8-hydroxy-2'-deoxy-guanosine measurement
Competitive ELISA assay for 8-OHdG was performed according to Schmerold & Niedermüller [21] using 8-OHdG-EIA kit (OXFORD, USA). Briefly, mitochondrial DNA was isolated from heart tissues using mtDNA Extractor CT Kit (Wako, USA) then 8-OHdG antibody and DNA sample were added to ELISA plate which has been precoated with 8-OHdG. The 8-OHdG in the sample competes with the 8-OHdG bound on the plate for the 8-OHdG antibody bites binding sties. The antibodies that are bound to the 8-OHdG in the sample were washed out of the well, while those that have bound to the 8-OHdG coated on the plate will remain. Following secondary antibody and chromogen, the color reaction was terminated and the absorbance was measured.
Mitochondrial Oxygen Consumption
The experimental procedures described are based on polarographic measurements of oxygen consumption by means of Clark-type oxygen electrode. For mitochondria studies the rate at which total chamber oxygen declines is referred to as the oxygen consumption rate [1].
Heart mitochondria were prepared by differential centrifugation using 250mM mannitol, 5 mM Na-HEPES (pH 7.0) and 0.5mM EGTA as the isolation medium [22]. Then, mitochondrial respiration was measured with a Clark-type oxygen electrode (Instech Laboratories Inc, Plymouth Meeting, Pa). Six isolated rat hearts from each group (both LETO and OLETF) were manually homogenized, using medium fitting glass-teflon Potter-Elvehjem homogenizer in isolation medium. The resultant homogenate was centrifuged twice and the mitochondria-enriched pellet was resuspended in respiration buffer (225 mM mannitol, 70mM sucrose, 10mM KH2PO4, and 1mM EGTA, pH 7.2). Oxygen consumption was measured in the presence of sequential administration of substrates (glutamate/malate for complex I, succinate for complex II, ascorbate for complex IV) was added in the following order and final concentration: 2.5mM glutamate, 2.5mM malate, 2mM adenosine diphosphate, 5mM succinate, and 1mM ascorbate. State (3) respiration was initiated by the addition of an excess of ADP (2mM). Respiration rates were expressed as micromoles of oxygen per minute per milligram of mitochondrial protein. Mitochondrial protein was determined according to the method of Lowry et al. [23]. Respiratory control ratio (RCR) was defined as the ratio of state 3 respiratory rate to state 4 rate. Isolation and assay of diabetic (OLETF) and control (LETO) mitochondria were always carried out at the same time as matched pairs.
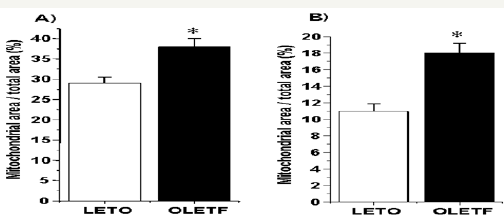
Figure 1: a) Mitochondrial size in LTEO (29±1.5) and OLETF (38±2) hearts. b) Mitochondrial size in LETO (11±0.9) and OLETF (18±1.2) thoracic aorta. Mitochondrial area was measured as percentage of total area on transmission electron microscope (TEM) micrographs
Results are means from 6 micrographs for each heart and 6 rats for each group. * P<0.05 compared with LETO rats.
Statistical Analysis
All results are expressed as means ± standard error of mean (SEM). Statistical analysis was performed by one-way analysis of variance (ANOVA, SuperAnova, Abacus Concepts; Berkeley, CA, USA). Differences between the control (LETO) and diabetes (OLETF) groups were assessed using the Student's t-test. Differences were considered significant at P < 0.05.
Results and Discussion
As the primary source of energy for the cardiac myocyte, mitochondria play a central role in cellular homeostasis. Not surprisingly, disruption of this critical organelle is regarded as a key contributor to the development of pathological states, including diabetic cardiomyopathy [24]. Mitochondria also compartmentalize reactions and molecules critical for metabolism, signaling, and programmed cell death [25]. Mitochondria are also dynamic, frequently changing size and shape and traveling long distances on cytoskeletal tracks. Sophisticated mechanisms that regulate different morphologies and distributions help to optimize mitochondrial function in response to changing intracellular needs and extracellular cues [26]. A better understanding of the mechanisms by which cardiomyocytes undergo apoptosis following diabetic oxidative stress would be helpful and may also provides additional targets for therapeutic intervention in the treatment of diabetic cardiomyopathy in the future. The electron microscopic examination of mitochondria from diabetic OLETF hearts in the present study revealed increases in both size and number (Figures 1 & 2). Similar findings were obtained by Shen et al. [27] who observed mitochondrial swelling in the OVE26 mouse model of diabetes, suggesting a compensatory mechanism against the enhanced apoptotic program. Moreover, Boudina et al. [28] demonstrated that diabetes and obesity are associated with mitochondrial proliferation in db/db heart without a coordinate increase in mitochondrial oxidative capacity and suggested that this mitochondrial biogenesis is an adaptive mechanism to overcome the decreased oxidative capacity.
Figure 2: a) Mitochondrial number in LTEO (21�1.6) and OLETF (32±1.2) hearts. b) Mitochondrial number in LETO (13±2) and OLETF (22±1.8) thoracic aorta. Mitochondrial number was counted and normalized to unit micrograph area. Results are means from 6 micrographs for each heart and 6 rats for each group.
* P<0.05 compared with LETO rats.

Figure 3: Fluorescence-activated cell sorting (FACScan) analysis of mitochondrial superoxide production.
a) Selection of mitochondria based on staining with Mito-Tracker Red and measurement of superoxide production based on the fluorescence of the oxidized H2DCF-DA by mitochondrial superoxide.
Significant increase in mitochondrial superoxide production expressed as mean fluorescence intensity in OLETF (883±22) compared with LETO (256±19) heart mitochondria.
* P<0.05 compared with LETO rats. Vertical bars indicate SE.

In the present study, the increased mitochondrial superoxide production in diabetic heart (Figure 3) coupled with increased mitochondrial DNA fragmentation and oxidation (Figures 4-6) imply that ROS defences in these diabetic hearts were not sufficiently increased to circumvent the increased oxidant burden.
Figure 4: The electrophoretic analysis of mtDNA from LETO and OLETF hearts is shown on a 1.5% agarose gel. Lane M, 100 bp DNA Ladder. Lane 1&2, mtDNA of LETO hearts and lane 3&4, mtDNA of OLETF hearts. The mtDNA of OLETF hearts are more fragmented than those of LETO hearts.
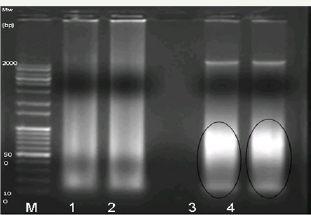
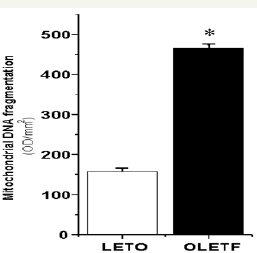
Figure 5: The density (expressed as OD x mm2) of mitochondrial DNA fragments in OLETF (465±11) and LETO (158±8.5) hearts.
* Significant variation between OLETF and LETO groups at p < 0.05.
Mitochondria appear to be at the crux of diabetic cardiac hypertrophy [9]. During oxidative phosphorylation, oxygen is reduced to water in four electron reductions at cytochrome oxidase (complex IV) in the mitochondrial ETC. More than 90% of the body's oxygen is consumed by the ETC [29]. It has been estimated that about 1-3% of the oxygen consumed in the mitochondria is released as superoxide and hydrogen peroxide (H2O2) at complexes I and III, making mitochondria a major source of endogenous reactive oxygen species (ROS) [30,31]. These ROS can damage macromolecules in mitochondria, including proteins, lipids, RNA, and DNA.
Figure 6: Mitochondrial DNA 8-OHdG content in LETO and OLETF hearts. The 8-OHdG level in the mtDNA of OLETF rat hearts (36.4±3.8) is significantly higher (2.9 fold increase) than that of control (LETO) hearts (12.5±2.1). Data are shown as means ± SEM. (n = 6).
* P<0.05 compared with LETO rats. Vertical bars indicate SE.

8-Hydroxy-2'-deoxyguanosine (8-OHdG) is one of the major products in oxidative DNA damage caused by reactive oxygen species [32]. The significant increase of 8-OHdG level in mitochondrial DNA of diabetic heart (Figure 6) indicated the oxidative damage of DNA leading to diabetic cardiomyopathy. This is where 8-OH-dG can be considered as a promutagenic lesion in DNA that is generated in response to a number of chemicals that induce oxidative stress [33] and it is widely used as a marker of oxidative injury [34].
Furthermore, Figures 4 & 5 showed that mitochondrial DNA is exhibited a maximal DNA fragmentation, a marker for apoptosis, in diabetic hearts when compared to control group. As mentioned by Sawyer & Van Houten [35] that in the last two decades, several studies have suggested that mtDNA is more susceptible than nuclear DNA to genotoxic agents, most notably ROS. This is augmented from the increased mitochondrial superoxide production (Figure 3) which could result in increased mitochondrial DNA oxidation as shown in Figure 6. In this context, numerous studies measured the levels of 8-OH-dG, a common byproduct of oxidative stress, both in mitochondrial and nuclear DNA (nDNA). Early reports indicated that the mtDNA had higher amounts of oxidative damage than the nuclear genome. Damage to the mtDNA, if not repaired, could lead to mutations during replications. More importantly, it could have further implications in cell physiology and, ultimately, in human health [36].
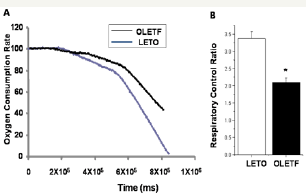
Mitochondrial electron transport chain (ETC) function can be profoundly affected by oxidative stress-associated pathologies, including diabetes mellitus because of its enhanced propensity for ROS-mediated damage. Our current data showed decrease in ETCrespiration measured by the alteration in respiratory control ratio (RCR) in diabetic OLETF heart mitochondria (Figure 7), which is in argment with other reports[37,38].
Figure 7: Impaired mitochondrial function in OLETF rat hearts.
a) Mitochondrial oxygen consumption rate in LETO and OLETF. b) Reduced respiratory control ratio in cardiac mitochondria from OLETF diabetic rats. Respiratory control ratio (RCR) was calculated as ratio of state 3 to state 4 rates. All results were normalized to mitochondrial protein content. Results are means from 6 animals for each group.
* P<0.05 compared with LETO rats. Vertical bars indicate SE.
Given a mitochondrial etiology for type II diabetes, the various stages in the progression of type II diabetes can be understood. The excessive reduction of the mitochondrial ETC electron carriers in the energy utilization tissues maximizes mitochondrial ROS production. The high serum insulin activates their Akt pathway, which phosphorylates the FOXOs. The departure of the FOXOs from the nucleus stops transcription of the stress response genes, including the mitochondrial antioxidant enzymes. It also suppresses PGC-1α transcription, which down regulates mitochondrial OXPHOS, further exacerbating the mitochondrial energy deficiency The resulting chronic mitochondrial oxidative stress erodes mitochondrial function and increases insulin resistance [15].
Moreover, our results are consistent with the recent findings reported by Dabkowski et al. [39] who observed significant decreases in oxygen consumption at complex I, III and complex IV in diabetic heart mitochondria. Furthermore, this is in agreement with previous reports of decreased respiration capacity in the diabetic heart mitochondria [40,41].
The impairment of cardiac mitochondrial function in OLETF rats in the current study is also consistent with the findings of [42-44] who demonstrated that cardiac mitochondria showed impairment in mitochondrial function and morphology, underscoring an important role for mitochondrial dysfunction in cardiac complications of diabetes.
References
- Murugan P, Pari L (2006) Antioxidant effect of tetrahydrocurcumin in streptozotocin-nicotinamide induced diabetic rats. Life Sciences 79(18): 1720-1728.
- Bell GI, Polonsky KS (2001) Diabetes mellitus and genetically programmed defects in ß-cell function. Nature 414(6865): 788-791.
- Kahn SE (2003) The relative contributions of insulin resistance and ß-cell dysfunction to the pathophysiology of Type 2 diabetes. Diabetologia 46(1): 3-19.
- Thomas RA, Dlugosz CK (2006) Tools and echniques for fighting the obesity epidemic: Program and abstracts of the American Pharmacists Association Annual Meeting and Exposition. San Francisco, USA.
- Shick SM, Wing RR, Klem ML, McGuire MT, Hill JO, et al. (1998) Persons successful at long-term weight loss and maintenance continue to consume a low calorie, low fat diet. J Am Dietetic Assoc 98(4): 408-413.LETO OLETF
- NHLB (National Heart, Lung, and Blood Institute), North American Association for the Study of Obesity (2006): The practical guide: identification, evaluation, and treatment of overweight and obesity in adults.Time (ms)
- Wallace DC, Brown MD, Lott MT (1996) Mitochondrial genetics, in Emery and Rimoin's Principles and Practice of Medical Genetics (Rimoin, DL, Reed Pyeritz, Bruce Korf, Eds.), Churchill Livingstone, London, UK, pp. 277-332.
- Kadowaki T, Sakura H, Otabe S, Yasuda K, Kadowaki H, et al. (1995) A subtype of diabetes mellitus associated with a mutation in the mitochondrial gene. Muscle Nerve Suppl 3: S137-S141.
- 9. Fein FS, Sonnenblick EH (1994) Diabetic cardiomyopathy. Cardiovasc Drugs Ther 8(1): 65-73.
- Koster HP, Hartog A, van Os CH, Bindels RJ, Koster JC (2000) Targeted overactivity of beta cell KATP channels induces profound neonatal diabetes. Cell 100(6): 645-654.
- Patti ME, Butte AJ, Crunkhorn S, Cusi K, Berria R, et al. (2003) Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: potential role of PGC1 and NRF1. Proc Natl Acad Sci USA 100(14): 8466-8471.
- Lee CH, Olson P, Evans RM (2003) Minireview: lipid metabolism, metabolic diseases, and peroxisome proliferator-activated receptors. Endocrinology 144(6): 2201-2207.
- Koutnikova H, Campuzano V, Foury F, Dolle P, Cazzalini O, et al. (1997) Studies of human, mouse and yeast homologues indicate a mitochondrial function for frataxin. Nat Genet 16(4): 345-351.
- Silva JP, Kohler M, Graff C, Oldfors A, Magnuson MA et al. (2000) Impaired insulin secretion and beta-cell loss in tissue-specific knockout mice with mitochondrial diabetes. Nat Genet 26(3): 336-340.
- Wallace DC (2001) Mouse models for mitochondrial disease. Am J Med Genet 106(1): 71-93.
- Rajapakse N, Shimizu K, Payne M, Busija D (2001) Isolation and characterization of intact mitochondria from neonatal rat brain. Brain Res Protoc 8(3): 176-183.
- Spallarossa P, Fabbi V, Manca S, Garibaldi G, Ghigliotti C, et al. (2005) Doxorubicin-induced expression of LOX-1 in H9c2 cardiac muscle cells and its role in apoptosis. Biochem Biophys Res Commun 335(1): 188196.
- Herrmann HM, Lorenz R, Voll M, Grünke W, Woith J, et al. (1994) A rapid and simple method for the isolation of apoptotic DNA fragments. Nucleic Acids Res 22(24): 5506-5507.
- Rudolf J, Wiesnera HS, Radovan Z (1991) Purification of mitochondrial DNA from total cellular DNA of small tissue samples. Gene 98(2): 277281.
- Sambrook J, Russel D (2001) Molecular Cloning: A Laboratory Manual, (3rd edn). Cold Spring Harbor Press, Cold Spring Harbor, NY, USA.
- Schmerold I, Niedermüller H (2001) Levels of 8-hydroxy-2'- deoxyguanosine in cellular DNA from 12 tissues of young and old Sprague-Dawley rats. Exp Gerontol 36(8): 1375-1386.
- Hofhaus G, Shakeley RM, Attardi G (1996) Use of polarography to detect respiration defects in cell cultures. Methods Enzymol 264: 476-483.
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the Folin phenol reagent. J Biol Chem 193(1): 265275.
- Rolo AP, Palmeira CM (2006) Diabetes and mitochondrial function: role of hyperglycemia and oxidative stress. Toxicol Appl Pharmacol 212(2): 167-178.
- Butow RA, Avadhani NG (2004) Mitochondrial signaling: the retrograde response. Mol Cell 14(1): 1-15.
- Bereiter-Hahn J, Voth M (1994) Dynamics of mitochondria in living cells: shape changes, dislocations, fusion, and fission of mitochondria. Microsc Res Tech 27(3): 198-219.
- Shen X, Zheng S, Thongboonkerd V, Xu M, Pierce WM, et al. (2004) Cardiac mitochondrial damage and biogenesis in a chronic model of type 1 diabetes. Am J Physiol Endocrinol Metab 287(5): E896-905.
- Boudina S, Sena S, Theobald H, Sheng X, Wright JJ, et al. (2007) Mitochondrial energetics in the heart in obesity-related diabetes: direct evidence for increased uncoupled respiration and activation of uncoupling proteins. Diabetes 56(10): 2457-2466.
- Boveris A, Oshino N, Chance B (1972) The cellular production of hydrogen peroxide. Biochem J 128(3): 617-630.
- Turrens JF, Boveris A (1980) Generation of superoxide anion by the NADH dehydrogenase of bovine heart mitochondria. Biochem J 191(2): 421-427.
- Wei YH, Scholes CP, King TE (1981) Ubisemiquinone radicals from the cytochrome b-c1 complex of mitochondrial electron transport chain: demonstration of QP-S radical formation. Biochem Biophys Res Commun 99(4): 1411-1419.
- Boiteux S, Radicella JP (1999) Base excision repair of 8-hydroxyguanine protects DNA from endogenous oxidative stress. Biochimie 81(1-2): 5967.
- Kasai H, Iwamoto-Tanaka N, Miyamoto T, Kawanami K, Kawanami S, et al. (2001) Life style and urinary 8-hydroxydeoxyguanosine, a marker of oxidative DNA damage: effects of exercise, working conditions, meat intake, body mass index, and smoking. Jpn J Cancer Res 92(1): 9-15.
- Toraason M (1999) 8-Hydroxyguanosine as a biomarker of workplace exposure. Biomarkers 4 (1): 3-26.
- Sawyer DE, Van Houten B (1999) Repair of DNA damage in mitochondria. Mutat Res 434: 161-176.
- Pettepher CC, LeDoux SP, Bohr VA, Wilson GL (1991) Repair of alkalilabile sites within the mitochondrial DNA of RINr 38 cells after exposure to the nitrosourea streptozotocin. J Biol Chem 266(5): 3113-3117.
- Kelley DE, He J, Menshikova EV, Ritov VB (2002) Dysfunction of mitochondria in human skeletal muscle in type 2 diabetes. Diabetes 51(10): 2944-2950.
- King KL, Young ME, Kerner J, Huang H, O'Shea KM, et al. (2007) Diabetes or peroxisome proliferator-activated receptor alpha agonist increases mitochondrial thioesterase I activity in heart. J Lipid Res 48(7): 15111517.
- Dabkowski ER, Williamson CL, Bukowski VC, Chapman RS, Leonard SS, et al. (2009) Diabetic cardiomyopathy-associated dysfunction in spatially distinct mitochondrial subpopulations. Am J Physiol Heart Circ Physiol 296(2): H359-369.
- Tomita M, Mukae S, Geshi E, Umetsu K, Nakatani M, et al. (1996) Mitochondrial respiratory impairment in streptozotocin-induced diabetic rat heart. Jpn Circ J 60(9): 673-682.
- Turko IV, Murad F (2003) Quantitative protein profiling in heart mitochondria from diabetic rats. J Biol Chem 278(37): 35844-35849.
- Bugger H, Chen D, Riehle C, Soto J, Theobald HA, et al. (2009) Tissue- specific remodeling of the mitochondrial proteome in type 1 diabetic akita mice. Diabetes 58(9): 1986-1997.
- Kanamori A, Tanaka K, Yajima Y (1995) Insulin sensitivity and mitochondrial gene mutation. Diabetes Care 18(2): 274-275.
- Wanders RJ, Van Roermund CW, Meijer AJ (1984) Analysis of the control of citrulline synthesis in isolated rat-liver mitochondria. Eur J Biochem 142(2): 247-254.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






