- Submissions

Full Text
Investigations in Gynecology Research & Womens Health
Quantification of Circulating Cell-Free DNA and its Clinical Applications in Cancer Management
Richard Jun Ma1,2, Aiguo Zhang1, Yunqing Ma1*, Wei Liu1*
1DiaCarta Inc, USA
2California North state University College of Medicine, USA
*Corresponding author:Yunqing Ma and Wei Liu, DiaCarta Inc, CA 94588, USA
Submission:April 30, 2024;Published: May 17, 2024

ISSN: 2577-2015 Volume4 Issue5
Abstract
As liquid biopsy gains more traction, quantification of the level of circulating cell-free DNA (cfDNA) in plasma has become an important biomarker to study in cancer diagnosis, tumor progression and treatment response monitoring in the whole process of cancer management. Here we summarized the commonly used plasma cfDNA quantification methods and reviewed cfDNA quantification in clinical applications in cancer management based on publications. The aim of the review is to provide scientific information and practical guidance for clinicians to choose the appropriate method for cfDNA quantitation despite the lack of standardization. We also hope to see the cfDNA quantification biomarker can be validated in larger clinical studies or clinical trials with different clinical indications in cancer management in the near future.
Introduction
Cell-free DNA (cfDNA) is released from dying or damaged cells into the body’s circulatory system. First discovered in plasma in 1948 by Mandel and Metais [1]), cfDNA were also discovered in other body fluids such as Cerebral Spinal Fluid (CSF) [2], urine [3,4], pleural fluid [5], saliva [6,7]. It is hypothesized that cfDNA is generated through both active (secretion) and passive (apoptosis in normal cells and necrosis in cancer cells) mechanisms [8]. In healthy individuals, most of the plasma cfDNA originates from the hematopoietic cells [9], while in cancer patients, tumor cells could release DNA, called circulating tumor DNA (ctDNA), into the blood stream as well as from the normal cells [10]. It is estimated that ctDNA accounts for a small fraction (0.01% to 1%) in early-stage cancer patients [11]. In advanced and metastatic cancer, the ctDNA fraction can go up to even 90% of the total cfDNA [12]. However, the increase of cfDNA in cancer patients is not totally from cancer cells. New studies indicate that the majority of the ctDNA is not from tumor cells, but from leukocytes in the blood [13]. More studies are needed to study the cfDNA origin in tumor cells to resolve the contradictions from the above observations.
Cell-free DNA are small pieces of DNA fragments in range of 120-220 bp with a maximum peak at 167 bp [14]. Unlike uniformly fragmented DNA released from apoptotic healthy cells, ctDNA released from tumor cells through necrosis varies in size [15]. Some researchers state that the ctDNA is around 50 to 100 bp [16]. This is supported by the evidence that enrichment of shorter fragments of DNA increases the ctDNA assay sensitivity [17]. In the human circulatory system, the cfDNA can exist in unbound/naked form or bound form internalized in extracellular vesicle (EVs)/exosomes [18].
cfDNA concentrations are found to be different between healthy/benign tumor patients and cancer patients [19]. The normal cfDNA concentration is normally in the range of 0 to 100ng/ml with an average of 30ng/ml whereas the cfDNA concentration ranges from 0 to 1000ng/ml with an average of 180ng/ml in cancer patients [20].
Accurate quantification of cfDNA is challenging at several levels. First of all, quantitation of cfDNA is sensitive to genomic DNA (gDNA) contamination derived from lysed blood cells in poorly manipulated samples [14], and it is critical to prevent hemolysis during processing and storage of cfDNA. Secondly, different DNA extraction methods showed varied DNA yield and fragment size distribution [18], causing underestimate of the real cfDNA concentration. Finally, varied DNA quantitation methods also give different cfDNA concentration [21], adding further variation. Hence, there is a need to standardize collection, handling, and preservation methods of blood, cfDNA isolation and cfDNA quantitation. In this review, we focused on the cfDNA quantitation area and summarized the commonly used cfDNA measurement methods from the literatures. With the comparison of these methods, we hope to provide comprehensive information and guidance for clinicians to choose the appropriate cfDNA quantification tools for their future clinical practice. We finally briefly reviewed the clinical utility and applications of cfDNA quantification as a biomarker.
Cell-free DNA Quantitation Methods
Direct and indirect quantification
The cfDNA quantitation methods include both direct quantitation and indirect quantitation methods. The direct method directly measures cfDNA concentration without DNA purification, such as direct PCR [22,23], direct SYBR Gold assay [23,24], and QuantiDNATM direct cfDNA test [25-27]. In indirect cfDNA quantitation method, the cfDNA needs to be purified from plasma or other body fluids and then quantitated with quantitative PCR (qPCR) [21,28-30], droplet digital PCR (ddPCR) [31], fluorescent dye (e.g. Qubit), or UV-spectrophotometry (e.g. NanoDrop). The advantages of the direct quantification methods are that they are quick and simple, but the drawback is the lack of accuracy due to matrix, reaction, or other interferences. The advantages of the indirect quantification methods lie in the elimination of all sources of interferences, but the major drawback is underestimation of cfDNA concentration due to sample loss during cfDNA extraction. The sample loss varies depending on the extraction methods used (column purification or magnetic beads purification) or the commercial vendors of the extraction kits. The cfDNA loss after extraction using QIAamp DNA Blood Mini Kit can be as high as 63.3% [22]. Unless a complex extraction control is included in extraction procedure to calibrate the loss, the cfDNA quantification after cfDNA extraction is unreliable. Although this cfDNA mis-quantification may not always affect the conclusion of clinical monitoring in a clinical study, it may have contributed to the contradictory results in some studies [32,33]. Inaccuracy and non- standardization of DNA quantification methods has also become the hurdle for the use of the cfDNA quantification in clinical practice [34]. These methods are summarized in (Table 1).
Table 1:Commonly used cfDNA quantitation methods.
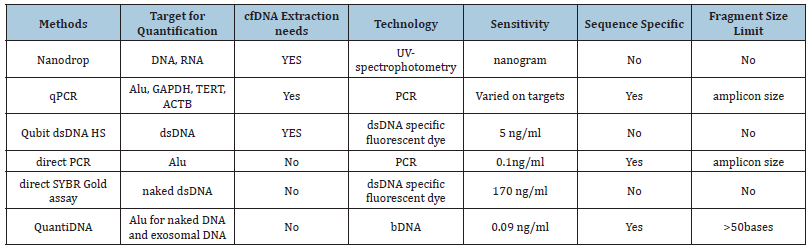
Nanodrop method
The common bench UV-spectrophotometric method such as NanoDrop cannot detect DNA below nanogram levels [3]. It can’t be used for direct measurement due to blood matrix interference. In addition, the UV-spectrophotometry cannot differentiate DNA from RNA and oligonucleotides, even single nucleotides. To measure the cfDNA quantitatively for healthy individuals, one to a few milliliters of plasma or other body fluids is needed to purify the cfDNA before quantification. Even after purification, the nanodrop method shows very poor correlation with other quantification methods such as ddPCR methods [32].
Qubit method
Fluorescent dye-based methods such as Qubit uses the double stranded specific dye such as SYBR gold, PicoGreen to measure the cfDNA concentration directly from plasma or purified DNA. The direct SYBR Gold assay quantitates cfDNA in microtiter plate directly from plasma or serum with Qubit Fluorometer. This method is simple and quick, but low sensitivity with a detection limit at only 170 ng/ml [25], which is far above the 0-100ng/ml of healthy individual [10]. In addition, in direct quantitation method, only freely unbound cfDNA is measured, as SYBR Green dye can only bind to free/naked DNA, not the DNA internalized in EV or exosomes [24]. However, Qubit quantification has been used for cfDNA quantification after DNA purification [32]. When compared to qPCR or ddPCR quantification methods, Qubit quantification method shows good correlation with these methods [32]. When a specific source of DNA is quantified from another, such as human cfDNA from mouse cfDNA, unlike the methods described below (e.g. qPCR, ddPCR or QuantiDNA™ methods), both Nanodrop and Qubit methods have limitations to quantitate species-specific cfDNA as they both detect DNA without sequence specificity.
qPCR and ddPCR methods
The qPCR method can also be used to quantitate cfDNA directly and indirectly. Breitbach et al developed a direct qPCR method with primers designed in a multi-locus L1PA2 sequence with loading 2ul of 1:40 diluted plasma [22]. Its LOQs were determined at 100 copies per reaction. It was reported that the majority of cfDNA in the blood of cancer patients was contained within exosomes, rather than floating freely DNA [18,35] Umetani et al. [15] developed another qPCR method with primers designed in ALU regions [23]. In order to measure the bound form of cfDNA, a proteinase K pretreatment was added. This ALU-qPCR method had a detection limit of 0.01pg of DNA which is equivalent to the plasma cfDNA concentration of 0.1ng/ml. Most of the qPCR quantification methods use purified cfDNA for quantification. These methods use either 1 million copies of conserved repeats such as ALU [23] or single copy housekeeping genes such as GAPDH [29], TERT [30], ACTB [21,31] as the reference gene to quantify.
A few parameters may affect the qPCR method accuracy. Single copy gene amplicon quantification may not always be the representative of the total amount of cfDNA [36], which may underestimate the total cfDNA in the clinical samples. qPCRbased cfDNA quantification is also highly variable according to the amplicon length [37]. Comparing with the total DNA amount gauged by PicoGreen, the DNA content determined by qPCR was severalfold lower [21]. This difference could be explained by the fact that PicoGreen or other dyes can detect nearly all DNA fragments while qPCR only quantitates amplifiable DNA.
ddPCR method is an absolute quantification tool for cfDNA copies, but the same principle of primer and amplicon design makes ddPCR carry the same drawbacks as qPCR does despite a more sensitive quantification method..
QuantiDNA™ direct cfDNA test and QuantiDNA™ DNA measurement assay
QuantiDNATM tests for cfDNA is highly sensitive and specific signal amplification nucleic acid probe assay based on the branched DNA (bDNA) technology [27] This assay is a direct hybridization assay without DNA/RNA extraction or PCR amplification. It uses microliters of plasma and detect 0.39ng/ml of cfDNA using the luminometer (QuantiDNATM DNA Measurement Assay) and 0.09ng/ ml cfDNA using Luminex MagPix (QuantiDNATM Direct cfDNA Test) after plasma dilution. This assay contains the proteinase K and detergents in its assay protocol therefore it can measure both the unbound freely cfDNA and protein/exosome bound cfDNA. The assay quantitates the cfDNA by hybridization with a seven oligos probeset covering a 200base ALU region [27]. The probeset has both CE (capturing the DNA targets to beads or plate well surface) and LE (hybridizing with bDNA molecules for signal amplification) types of oligos where any two neighboring CE, LE oligo can quantitate DNA fragments over 50 bases.
QuantiDNATM DNA Measurement Assay, have shown the accuracy of quantification with 5 to 10% variation from the standards (DiaCarta, unpublished), exceeding the accuracy of the methods that requires cfDNA extraction, that may underestimate the actual cfDNA DNA by 40 to 60%. This quantification method only requires 10 to 20 microliters of plasma and is currently used in many clinical applications [26,28,38] [Table 1].
In summary, there is a need for method standardization of cfDNA quantification before the clinical studies can be carefully compared and solid conclusion is generated. Although Qubit and qPCR are often used by researchers, many of them have not included an extraction control to calibrate the accuracy of the quantification methods. It is necessary to evaluate QuantiDNATM cfDNA quantification methods side by side with the traditional methods before picking a reliable cfDNA quantification method for clinical applications.
cfDNA Quantification in Clinical Applications of Cancer Management
Many studies have indicated that the cfDNA levels are much higher in cancer patients than in healthy controls, and patients in late-stage cancer than in early-stage cancer [39], indicating the role of cfDNA levels as a biomarker for cancer prognosis. Fan et al. reported that plasma cfDNA concentration was markedly higher among preoperative lung cancer patients when compared to healthy subjects [40] Austin et al. measured the cfDNA concentrations in 178 patients with colon, pancreatic, lung or ovarian cancer and 64 healthy individuals and found the levels of cfDNA among the cancer patients are substantially higher than healthy subjects [13]. In breast cancer, the level of cfDNA is significantly higher in cancer patients than in healthy people and people with benign tumors [41,42], higher in metastatic patients than in progressionfree patients [42,43]. Although the cfDNA quantification marker has better sensitivity and specificity than traditional protein biomarkers such as CEA and CA (cancer antigen), the cfDNA is less than ideal to be used as a diagnostic marker.
In management of different types of cancers, cfDNA quantitation can also be used for response monitoring of treatments of various types, including radiotherapy, chemotherapy, target therapy, and immunotherapy [44-47]. cfDNA level changes in breast cancer patients is correlated with chemotherapy response [48,49]. When compared with the traditional biomarkers such as CA 15-3 and alkaline phosphatase, total cfDNA is a better predictor for both treatment response and overall survival [50]. The cfDNA level is also higher in recurrent breast cancer patients than in non-recurrent patients [51]. These studies have shown that cfDNA concentrations may have a great prognostic value [14,52]. In Ovarian Cancer (OC), cfDNA quantification has been demonstrated to be an effective biomarker for treatment (e.g. chemotherapy) response in OC [53,54] and predicts disease-specific survival in OC [55,56].
There are recent reviews on cfDNA quantification and ctDNA analysis [35,56] and their applications in clinical review. Here, we will not discuss the ctDNA analyses (e.g. mutation analysis and methylation analysis) that are used for cancer screening, Molecular Residual Disease (MRD) detection for recurrence prediction, or treatment responses. Although ctDNA is tumor-specific, they are more time consuming and costly, and hard to be applied for frequent monitoring compared to cfDNA quantification monitoring. The two types of analysis can complement each other and provide more comprehensive information in the whole cancer management process.
Conclusion
Current clinical studies use qPCR and Qubit methods for cfDNA quantification by ignoring the fact that these methods underestimate cfDNA level. This may cause wrong conclusions when studying the correlation of cfDNA with cancer progression and treatment responses. QuantiDNATM direct cfDNA quantitation and other methods should also be evaluated in clinical studies side by side with the traditional methods. After all, it is critical to have a simple and reliable cfDNA quantification method if the biomarker is to be used in clinical settings.
References
- Mandel P, and Metais P (1948) Nuclear acids in human blood plasma. C R Seances Soc Biol Fil 142(3-4): 241-243.
- Rhodes CH, Honsinger C, Sorenson GD (1994) Detection of tumor-derived DNA in cerebrospinal fluid. J Neuropathol Exp Neurol 53(4): 364-368.
- Sidransky D, Eschenbach AV, Tsai YC, Jones P, Summerhayes I, et al. (1991) Identification of p53 gene mutations in bladder cancers and urine samples. Science 252(5006): 706-709.
- Zhang J, Tong KL, Li PK, Chan AY, Yeung CK, et al. (1999) Presence of donor-and recipient-derived DNA in cell-free urine samples of renal transplantation recipients: urinary DNA chimerism. Clin Chem 45(10): 1741-1746.
- Sriram KB, Relan V, Clarke BE, Duhig EE, Windsor MN, et al. (2012) Pleural fluid cell-free DNA integrity index to identify cytologically negative malignant pleural effusions including mesotheliomas. BMC Cancer 12: 428.
- Mithani SK, Smith IM, Zhou S, Gray A, Koch WM, et al. (2007) Mitochondrial resequencing arrays detect tumor-specific mutations in salivary rinses of patients with head and neck cancer. Clin Cancer Res 13(24): 7335-7340.
- Wang Y, Springer S, Mulvey CL, Silliman N, Schaefer J, et al. (2015) Detection of somatic mutations and HPV in the saliva and plasma of patients with head and neck squamous cell carcinomas. Sci Transl Med 7(293): 293ra104.
- Stejskal P, Goodarzi H, Srovnal J, Hajdúch M, van ’t Veer LJ, et al. (2023) Circulating tumor nucleic acids: biology, release mechanisms, and clinical relevance. Molecular Cancer 22(1):15.
- Lui YY, Chik KW, Chiu RWK, Ho CY, Lam CWK, et al. (2002) Predominant hematopoietic origin of cell-free DNA in plasma and serum after sex-mismatched bone marrow transplantation. Clin Chem 48(3): 421-427.
- Yan Y, Guo Q, Wang FH, Adhikari R, Zhu ZY, et al. (2021) Cell-free DNA: hope and potential application in cancer. Front Cell Dev Biol 9: 639233.
- Parsons HA, Beaver JA, Cimino-Mathews A, Ali SM, Axilbund J, et al. (2017) Individualized molecular analyses guide efforts (IMAGE): a prospective study of molecular profiling of tissue and blood in metastatic triple-negative breast cancer. Clin Cancer Res 23(2): 379-386.
- Elazezy M, Joosse SA (2018) Techniques of using circulating tumor DNA as a liquid biopsy component in cancer management. Comput Struct Biotechnol J 16: 370-378.
- Mattox AK, Douville C, Wang Y, Popoli M, Ptak J, et al. (2023) The origin of highly elevated cell-free DNA in healthy individuals and patients with pancreatic, colorectal, lung, or ovarian cancer. Cancer Discov 13(10): 2166-2179.
- Alcaide M, Cheung M, Hillman J, Rassekh SR, Deyell RJ, et al. (2020) Evaluating the quantity, quality and size distribution of cell-free DNA by multiplex droplet digital PCR. Scientific Reports 10(1): 12564.
- Umetani N, Kim J, Hiramatsu S, Reber HA, Hines OJ, et al. (2006) Increased integrity of free circulating DNA in sera of patients with colorectal or periampullary cancer: direct quantitative PCR for ALU repeats. Clinical Chemistry 52(6): 1062-1069.
- Jiang P, Chan CWM, Chan KCA, Cheng SH, Wong J, et al. (2015) Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc Natl Acad Sci USA 112(11): E1317-1325.
- Liu Y, Liu Y, Wang Y, Li L, Yao W, et al. (2021) Increased detection of circulating tumor DNA by short fragment enrichment. Transl Lung Cancer Res 10(3): 1501-1511.
- Grabuschnig S, Bronkhorst AJ, Holdenrieder S, Rodriguez IR, Schliep KP (2020) Putative origins of cell-free DNA in humans: a review of active and passive nucleic acid release mechanisms. Int J Mol Sci 21(21): 8062.
- Koukourakis MI, Xanthopoulou E, Koukourakis IM, Fortis SP, Kesesidis N, et al. (2023) circulating plasma cell-free DNA (cfDNA) as a predictive biomarker for radiotherapy: results from a prospective trial in head and neck cancer. Cancer Diagn Progn 3(5): 551-557.
- George NG, Rishi B, Singh A, Vishmaya S, Kumar R, et al. (2024) Early prognosis prediction in acute myeloid and acute lymphoid leukemia patients using cell-free DNA concentration ratios. Front Mol Biosci 10: 1333943.
- Szpechcinski A, Struniawska R, Zaleska J, Chabowski M, Orlowski T, et al. (2008) Evaluation of fluorescence-based methods for total vs. amplifiable DNA quantification in plasma of lung cancer patients. J Physiol Pharmacol 6: 675-681.
- Breitbach S, Tug S, Helmig S, Zahn D, Kkubiak T (2014) direct quantification of cell-free, circulating DNA from unpurified plasma. PloS ONE 9(3): e87838
- Panagopoulou M, Esteller M, Chatzaki E (2021) Circulating cell-free DNA in breast cancer: searching for hidden information towards precision medicine. Cancers (Basel) 13(4): 728.
- Goldshtein H, Hausmann MJ, Douvdevani A (2009) A rapid direct fluorescent assay for cell-free DNA quantification in biological fluids. Ann Clin Biochem 46(Pt 6):488-494.
- Zhong Y, Fan Q, Zhou Z, Wang Y, He K, et al. (2020) Plasma cfDNA as a potential biomarker to evaluate the efficacy of chemotherapy in gastric cancer. Cancer Manag Res 12: 3099-3106.
- https://patents.google.com/patent/US8404444B2/en
- Shao X, He Y, Ji M, Chen X, Qi J, et al. (2015) Quantitative analysis of cell-free DNA in ovarian cancer. Oncology Letters 10: 3478-3482.
- Zanetti-Dällenbach RA, Schmid S, Wight E, Holzgreve W, Ladewig A, et (2007) Levels of circulating cell-free serum DNA in benign and malignant breast lesions. The International Journal of Biological Markers 22(2): 95-99.
- Hashad D, Sorour A, Ghazal A, Talaat I (2012) Free circulating tumor DNA as a diagnostic marker for breast cancer. J Clin Lab Anal 26(6): 467-472.
- Azzoli CG, Park S, Gomez J, Krug L, Miller V, et al. (2005) Measurements of total DNA and methylated tumor suppressor genes in the plasma of patients with metastatic non-small cell lung cancer (NSCLC) before and after chemotherapy, as potential biomarkers for response to treatment. J Clin Oncol 23:7203.
- Van Ginkel JH, van den Broek DA, van Kuik J, Linders D, De Weger R, et al. (2017) Preanalytical blood sample workup for cell-free DNA analysis using Droplet Digital PCR for future molecular cancer diagnostics. Cancer Med 6(10): 2297-2307.
- Kumar S, Guleria R, Singh V, Bharti AC, Mohan A, et al. (2010) Plasma DNA level in predicting therapeutic efficacy in advanced nonsmall cell lung cancer. Eur Respir J 36(4): 885-892.
- Pan S, Xia W, Ding Q, Shu Y, Xu T, et al. (2012) Can plasma DNA monitoring be employed in personalized chemotherapy for patients with advanced lung cancer? Biomed Pharmacother 66(2): 131-137.
- Dao J, Conway PJ, Subramani B, Meyyappan D, Russell S, et al. (2023) using cfDNA and ctDNA as oncologic markers: a path to clinical validation. Int J Mol Sci 24(17): 13219.
- Kahlert C, Melo SA, Protopopov A, Tang J, Seth S, et al. (2014) Identification of double-stranded genomic DNA spanning all chromosomes with mutated kras and p53 dna in the serum exosomes of patients with pancreatic cancer. J Biol Chem 289(7): 3869-3875.
- Bronkhorst AJ, Ungerer V, Holdenrieder S (2019) Comparison of methods for the quantification of cell-free DNA isolated from cell culture supernatant. Tumor Biology 41(8): 1010428319866369.
- Mouliere F, Robert B, Peyrotte EA, Rio MD, Ychou M, et al. (2011) High fragmentation characterizes tumour-derived circulating DNA. PloS ONE 6(9): e23418.
- Lockney NA, Randal Henderson R, Swarts SG, Zhang Z, Zhang B, et al. (2020) Circulating cell-free DNA correlates with body integral dose and radiation modality in prostate cancer. Int J Particle Ther 7(2): 21-30.
- Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, et al. (2014) Detection of circulating tumor DNA in early-and late-stage human malignancies. Sci Transl Med 6(224): 224ra24.
- Fan Y, Shi M, Chen S, Ju G, Chen L, et al. (2019) Analysis of serum cfDNA concentration and integrity before and after surgery in patients with lung cancer. Cell Mol Biol (Noisy-Le-Grand) 65(6): 56-63.
- Miao Y, Fan Y, Zhang L, Ma T, Li R (2009) Clinical value of plasma cfDNA concentration and integrity in breast cancer patients. Cell Mol Biol (Noisy-Le-Grand) 65(6): 64-72.
- Stebbing J, Takis PG, Sands CJ, Maslen L, Lewis MR, et al. (2023) Comparison of phenomics and cfDNA in a large breast screening population: the Breast Screening and Monitoring Study (BSMS). Oncogene 42(11):825-832.
- Ibrahim IH, Kamel MM, Ghareeb M (2016) Circulating DNA in Egyptian Women with Breast Cancer. Asian Pac J Cancer Prev 17(6): 2989-2993.
- Gristina V, Barraco N, Mantia ML, Castellana L, Insalaco L, et al. (2022) Clinical potential of circulating cell-free DNA (cfDNA) for longitudinally monitoring clinical outcomes in the first- line setting of non-small-cell lung cancer (NSCLC): A real-world prospective study. Cancers (Basel) 14(23): 6013.
- Zhou X, Li C, Zhang Z, Li DY, Du J, et al. (2021) Kinetics of plasma cfDNA predicts clinical response in non-small cell lung cancer patients. Scientific Reports 11(1): 7633.
- Peng W, Liu Y, Sha H, Wen S, Fang Y, et al. (2023) Relationship between plasma circulating cell-free DNA concentration and treatment outcomes including prognosis in patients with advanced non-small cell lung cancer. BMC Pulmonary Medicine 23(1): 348.
- Peled M, Agassi R, Czeiger D, Ariad S, Riff R, et al. (2020) Cell-free DNA concentration in patients with clinical or mammographic suspicion of breast cancer. Scientific Reports 10(1): 14601.
- Ma G, Wang J, Huang H, Han X, Xu J, et al. (2020) Identification of the plasma total cfDNA level before and after chemotherapy as an indicator of the neoadjuvant chemotherapy response in locally advanced breast cancer. Cancer Med 9(7): 2271-2282.
- Wang W, Zhang W, Su L, Sang J, Wang S, et al. (2019) Plasma cell-free DNA integrity: a potential biomarker to monitor the response of breast cancer to neoadjuvant chemotherapy. Transl Cancer Res 8(4): 1531-1539.
- Fernandez-Garcia D, Hills A, Page K, Hastings RK, Toghill B, et al. (2019) Plasma cell-free DNA (cfDNA) as a predictive and prognostic marker in patients with metastatic breast cancer. Breast Cancer Research 21(1): 149.
- Bera A, Russ E, Karaian J, Landa A, Radhakrishnan S, et al. (2022) Circulating cell-free DNA in serum as a marker for the early detection of tumor recurrence in breast cancer patients. Cancer Diagn Progn 2(3): 285-292.
- Volik S, Alcaide M, Morin RD, Collins CC (2016) Cell-free DNA (cfDNA): clinical significance and utility in cancer shaped by emerging technologies. Mol Cancer Res 14(10): 898-908.
- Cappizi E, Gabusi E, Grigioni AD, Iaco PD, Rosati M, et al. (2008) Quantification of free plasma DNA before and after chemotherapy in patients with advanced epithelial ovarian cancer. Diagnostic Molecular Pathology 17(1): 34-38.
- Steffensen KD, Madsen CV, Andersen RF, Waldstrøm M, Adimi P, et al. (2014) Prognostic importance of cell-free DNA in chemotherapy resistant ovarian cancer treated with bevacizumab. Eur J Cancer 50(15): 2611-2618.
- Kamat AA, Baldwin M, Urbauer D, Dang D, Han LY, et al. (2010) Plasma cell-free DNA in ovarian cancer: An independent prognostic biomarker. Cancer 116: 1918-1925.
- Telekes A, Horváth A (2022) The role of cell-free DNA in cancer treatment decision making. Cancers (Basel) 14(24): 6115.
© 2024 Yunqing Ma and Wei Liu. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






