- Submissions

Full Text
Gastroenterology Medicine & Research
Bacterial Community Composition and its Functional Potential in Ulcerative Colitis Patients: A Case Study
Stine Karstenskov Østergaard1, Christina Karmisholt Overgaard1, Nadieh de Jonge1, Lone Larsen2,3 and Jeppe Lund Nielsen1*
1Department of Chemistry and Bioscience, Aalborg University, Denmark
2Department of Gastroenterology and Hepatology, Aalborg University Hospital, Aalborg, Denmark
3Center for Molecular Prediction of Inflammatory Bowel Disease, PREDICT, Department of Clinical Medicine, The Faculty of Medicine, Aalborg University, Aalborg, Denmark
*Corresponding author:Jeppe Lund Nielsen, Department of Chemistry and Bioscience, Aalborg University, Fredrik Bajers vej 7H, 9220 Aalborg East, Denmark
Submission:July 12, 2023;Published: July 24, 2023

ISSN 2637-7632Volume7 Issue3
Abstract
Ulcerative Colitis (UC) is a chronic inflammatory bowel disease characterized by recurring inflammation in the colon. This study aimed to showcase the challenges related to the characterization of the enteric microbial community structure using 16S rRNA gene amplicon sequencing and its functional potential using metagenomic sequencing in five patients with UC during active disease and remission. The results revealed inter-individual and intra-individual differences in the microbial community composition. Differential abundance analysis identified specific genera associated with disease state, such as Faecalibacterium and Anaerostipes, which showed positive- and negative correlations, respectively. Prevotella was observed only during active disease. The high level of inter-individual taxonomic differences makes it difficult to link the changes to the disease. Functional analysis identified genes related to virulence and inflammatory bowel disease specifically during active disease. Although the approach showed great potential, it was limited by the vast amount of sequencing effort used on host DNA. Further research with a larger cohort and optimized DNA extraction protocols is needed in order validate the results and explore the functional roles of relevant epithelial-associated bacteria which is essential for unravelling the intricate host-microbiota interactions underlying disease pathogenesis.
Abbreviations:UC: Ulcerative Colitis; SCFA: Short-Chain Fatty Acids; IM: Inner Membrane; OM: Outer Membrane
Introduction
Ulcerative Colitis (UC) is a chronic inflammatory bowel disease in which the pathogenesis is thought to arise from a complex interplay between environmental factors, the microbiome and immune dysregulation in genetically susceptible individuals [1,2]. Conventional treatments for UC often have limited efficacy and unwanted side effects, necessitating a deeper understanding of the underlying mechanisms. It is currently a challenge to noninvasively diagnose and assess disease activity due to the lack of markers specific for UC [3]. As such it is highly relevant to search for more reliable biomarkers. It is known that environmental exposures such as diet, smoking, hygiene, antibiotics, mode of birth (vaginal vs. cesarean section) and breast feeding modulates the intestinal microbiome, and they are all considered to be risk factors for developing UC [4,5]. There seem to be consensus that changes in enteric microbiome is closely related to UC pathogenesis [6]. Several studies have documented compositional changes of the intestinal microbiota between patients with UC and healthy individuals, especially regarding microbial diversity and relative abundance of specific bacteria [7-9]. The studies have aimed to link these taxonomic differences to the disease but there is a lack of consensus in literature as to whether some species correlate positively or negatively. Most of these studies are limited by low taxonomic resolution, and furthermore lack correlation to function and inflammatory markers (e.g., calprotectin cytokines) [7-9]. Combined with the fact that inter-individual differences in the microbial community composition account ~50% of the variation [10] it is difficult to exclusively link the changed abundances to UC. Due to this complexity, focus has moved from taxonomic profiling to functional profiling [11-13]. This case study focuses on the characterization of the gut microbial community structure and function of biopsy samples in UC patients. By investigating differences between disease states and exploring the functional potential of the microbiome, we aim to shed light on methods suitable to investigate disease pathogenesis and find new potential therapeutic targets.
Methods
Ethical declaration and patient demographics
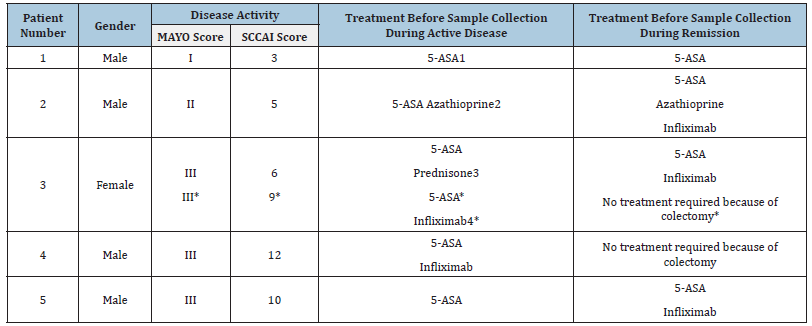
The project was approved by The North Denmark Region Committee on Health Research Ethics (N-20180043) and included five patients with Ulcerative Colitis (UC) from Aalborg University Hospital (Table 1). Inclusion criteria: UC diagnosis and >18 years of age. Exclusion criterium: former colectomy. One biopsy from the sigmoid colon and one biopsy from the rectum were collected both during active disease and during remission. Disease activity was confirmed and assessed with sigmoidoscopy using the MAYO and SCCAI score.
Table 1:Demographic and clinical characteristics. Note that the MAYO score only reflects endoscopic findings.
1. 5-aminosalicylic acid (5-ASA) is an anti-inflammatory drug (Garud and Peppercorn, 2009)
2. Immunosuppressant drug (Garud and Peppercorn, 2009)
3. Corticosteroids (Garud and Peppercorn, 2009)
4. Biological treatment consistent of monoclonal antibodies that binds the pro-inflammatory cytokine TNF-α
(Garud and Peppercorn, 2009; Aggarwal et al. 2017)
*This sample was taken in connection with the colectomy of patient 3.
DNA-extraction, amplicon preparation, sequencing, and analysis
Total genomic DNA was extracted using the DNeasy® Blood & Tissue kit (Qiagen) in accordance with specifications of the manufacturer. The V1-V3 region of the 16S rRNA gene [14] was amplified and sequenced on a MiSeq (Illumina, USA) using MiSeq reagent kit V3 (2 x 300 PE). The raw sequencing data was summarized into amplicon sequencing variants (ASVs) using AmpProc v. 5.1.0. beta 2.8 (https://github.com/eyashiro/AmpProc) using the USEARCH v. 11.0.667 [15] workflow in paired end mode. Taxonomic classification was performed in QIIME using SILVA release S132 as a reference database [16]. The resulting ASVs were analyzed in R v. 4.0.2 [17] through Rstudio v. 4.0.0.28 [18] using the ampvis2 v. 2.6.5 [19] and ggplot2 package [20].
Shallow metagenomic sequencing and analysis
The sigmoid colon biopsy specimens from patient 3 and the rectum biopsy specimen from patient 5 during both active disease and remission were sequenced on an Illumina MiSeq platform using reagent kit v3 (2x300 PE) (Illumina, USA). To separate reads of human and bacterial origin the trimmed reads were mapped against the RefSeq reference genome: Homo sapiens genome GRCh38 (accession number: PRJNA168 and PRJNA31257). The unmapped reads were assembled using default settings according to patient and disease state resulting in four assemblies. A Pfam domain search of the translated contig lists were done using the Pfam-A v. 33.1 database.
Data availability
Metagenomic and 16S rRNA gene amplicon data is available at the European Nucleotide Archive (ENA) under project accession number PRJEB64262 and will be made public upon publication.
Results
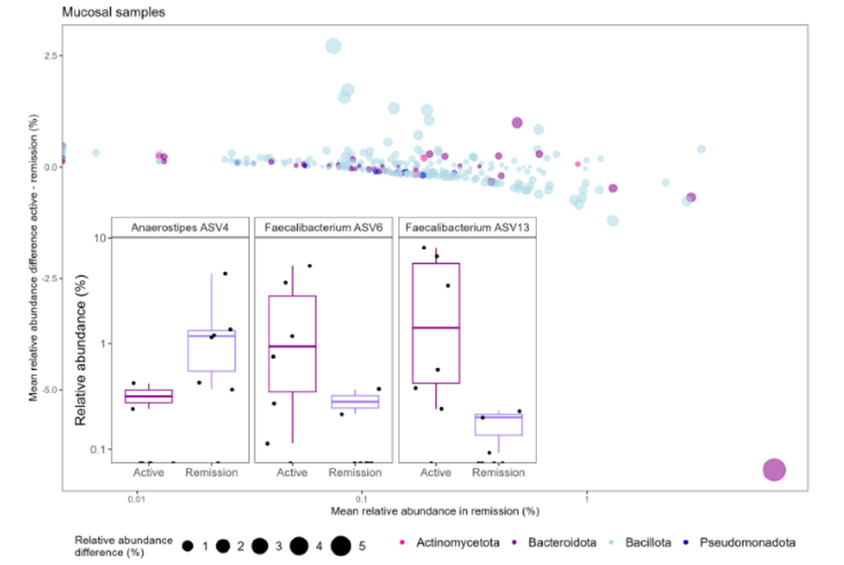
The alpha diversity analysis revealed no significant differences in richness between disease states (data not shown). The taxonomic composition analysis identified inter-individual differences, with Bacillota (49.6-92.9%), Bacteriodota (3.8-43.7 %), Actinomycetota (0.5-3.8 %) and Pseudomonadota (0-5.8 %) being the predominant phyla in all samples. Intra-individual variations were observed between disease states, particularly in the abundance of Faecalibacterium which increased during active disease while Anaerostipes decreased (Figure 1). To determine whether some bacteria were differentially abundant between disease states, a plot comparing the mean relative abundance in remission to the difference in mean relative abundance between disease states (Active - Remission) was created. Two sequencing variants belonging to the genus Faecalibacterium (ASV6 (p=0.01) and ASV13(p=0.03) and Anaerostipes ASV4 (p=0.01) had a mean relative abundance difference of ≥ 1 % between disease states.
Figure 1:Differentially abundant bacteria according to disease state. The mean relative abundance difference in percent (Active-Remission) is plotted against the mean relative abundance in percent during remission. The x-axis is logarithmic. Each dot represents an ASV. Color depicts to which phylum the bacteria belonged. Size depicts the difference from mean relative abundance during remission in percent. Bacteria were considered differentially abundant between disease states if the mean relative abundance difference were ≥ 1 %. The bacteria having a mean relative abundance difference > 0 were more abundant during active disease and those having a mean relative abundance difference < 0 were more abundant during remission. Boxplot displaying the three bacteria that had mean relative abundance difference > 1 % and p < 0.05. P-values were calculated using the Wilcoxon rank sum test and indicates whether or not there were a significant difference in abundance of the bacteria between the two disease states. Each dot represents a sample and color depicts disease state. The y-axis is logarithmic.
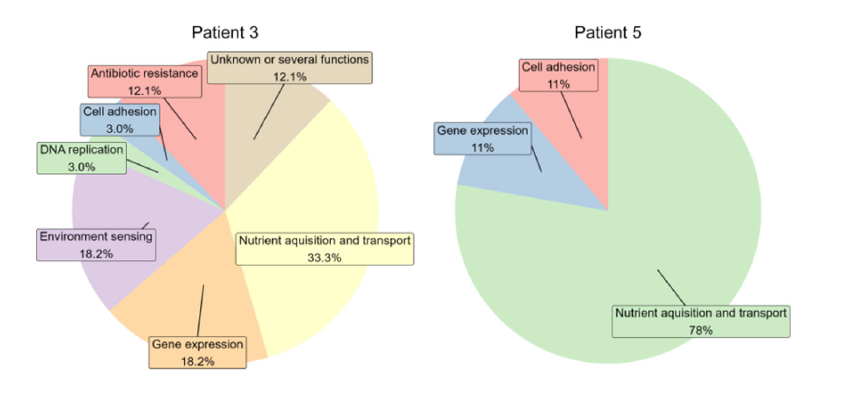
Functional analysis using shotgun metagenomic sequencing showed that more genes were identified and annotated during active disease compared to remission in both patients, although the amount of non-human reads was generally low (Table 2). Differences in the functional potential between disease states were observed. Genes related to virulence and/or UC were detected exclusively during active disease including the plug domain of TonB-dependent transport system (PF07715.12) and ABC_trans (PF00005.28) which are both Inner Membrane (IM) associated transporters. Other parts of the TonB-dependent transport system were also present in both patients including the outer membrane (OM) associated transporter TonB_dep_rec (PF00593.25) in patient 3 and the OM-associated TonB_C (PF03544.15) in patient 5. Increased potential for cell adhesion and antibiotic resistancerelated proteins was observed during active disease (Figure 2).
Table 2:Shotgun metagenomic sequencing data, mapping, and assembly results.
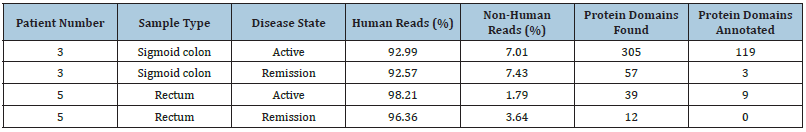
Figure 2:Functional categories of relevant protein domains during active disease. Pie charts were constructed for each patient. Colors depicts functional categories.
Discussion
Despite no significant differences in alpha diversity, variations in taxonomic composition were observed between disease states, suggesting a dynamic nature of the microbiome during active disease and remission. The complexity of dysbiosis in UC was evident in the inter-individual and intra-individual variations at the genus level. Faecalibacterium, known for its SCFA production [21], exhibited conflicting abundance changes during active disease. Anaerostipes, inhabiting various SCFA metabolisms [22], showed a consistent decrease during active disease, potentially contributing to reduced SCFA availability in UC patients. Furthermore, the presence of Prevotella [23,24], a potential pathobiont, during active disease raises intriguing possibilities regarding its role in UC pathogenesis. Although differences between disease states was observed in this small cohort size, it was difficult to exclusively link the changed abundancies to the disease.
The functional analysis uncovered genes related to virulence and antibiotic resistance during active disease, emphasizing the importance of considering not only taxonomic composition but also functional characteristics of the microbiome. The TonB-dependent transport system facilitates transport of nutrients across the OM in Gram-negative bacteria, but it has been shown that mutations in TonB in several bacterial species resulted in a loss of virulence in animal models [25]. This suggest that some pathobionts and pathogens use the ability to acquire certain substrates in initiation and establishment of an infection. The low number of annotated genes limited the analysis, and we hypothesize that this is likely attributed to the vast amount of sequencing effort used on host DNA.
This case study emphasizes the challenge of identifying reliable biomarkers in Ulcerative Colitis (UC) and gastrointestinal diseases. Understanding the significance of epithelial-associated bacteria is crucial for unraveling complex host-microbiota interactions. Based on this study, we suggest to improve and optimize the DNA extraction protocol for biopsies, aiming at increasing the bacterialto- human DNA ratio. One potential optimization step involves the inclusion of host depletion, which can enhance the microbial signal in both 16S rRNA gene amplicon sequencing as well as metagenomic sequencing. An increased taxonomic resolution can be obtained by selecting a sequencing platform capable of long read sequencing. Additionally, comparing inflamed and non-inflamed tissue within the same patient and utilizing large cohorts considering the dynamic nature of the microbiome influenced by factors like diet is advisable. This comprehensive approach, coupled with a holistic omics strategy, will provide a deeper understanding of the disease and its associated microbial changes.
References
- Lane ER, Zisman TL, Suskind DL (2017) The microbiota in inflammatory bowel disease: Current and therapeutic insights. J Inflamm Res 10: 63-73.
- Pickard JM, Zeng MY, Caruso R, Núñez G (2017) Gut microbiota: Role in pathogen colonization, immune responses, and inflammatory disease. Immunol Rev 279(1): 70-89.
- Guo X, Huang C, Xu J, Xu H, Liu L, et al. (2022) Gut microbiota is a potential biomarker in inflammatory bowel disease. Front Nutr 8: 818902.
- Ananthakrishnan AN (2015) Epidemiology and risk factors for IBD. Nat Rev Gastroenterol Hepatol 12(4): 205-217.
- Mueller NT, Bakacs E, Combellick J, Grigoryan Z, Dominguez-Bello MG (2015) The infant microbiome development: Mom matters. Trends Mol Med 21(2): 109-117.
- Schirmer M, Garner A, Vlamakis H, Xavier RJ (2019) Microbial genes and pathways in inflammatory bowel disease. Nat Rev Microbiol 17(8): 497-511.
- Saitoh S, Noda S, Aiba Y, Takagi A, Sakamoto M, et al. (2002) Bacteroides ovatus as the predominant commensal intestinal microbe causing a systemic antibody response in inflammatory bowel disease. Clin Diagn Lab Immunol 9(1): 54-59.
- Frank DN, St. Amand AL, Feldman RA, Boedeker EC, Harpaz N, et al. (2007) Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A 104(34): 13780-13785.
- Bajer L, Kverka M, Kostovcik M, Macinga P, Dvorak J, et al. (2017) Distinct gut microbiota profiles in patients with primary sclerosing cholangitis and ulcerative colitis. World J Gastroenterol 23(25): 4548-4558.
- Schirmer M, Denson L, Vlamakis H, Franzosa EA, Thomas S, Gotman NM, et al. (2018) Compositional and temporal changes in the Gut microbiome of pediatric ulcerative colitis patients are linked to disease course. Cell Host Microbe 24(4): 600-610.e4.
- Bajer L, Kverka M, Kostovcik M, Macinga P, Dvorak J, et al. (2017) Distinct gut microbiota profiles in patients with primary sclerosing cholangitis and ulcerative colitis. World J Gastroenterol 23(25): 4548-4558.
- Lee JG, Han DS, Jo SV, Reum Lee A, Park CH, et al. (2019) Characteristics and pathogenic role of adherent-invasive Escherichia coli in inflammatory bowel disease: Potential impact on clinical outcomes. PLoS One 14(4): e0216165.
- Moustafa A, Li W, Anderson EL, Wong EHM, Dulai PS, et al. (2018) Genetic risk, dysbiosis, and treatment stratification using host genome and gut microbiome in inflammatory bowel disease. Clin Transl Gastroenterol 9(1): e132.
- Human Microbiome Project Consortium (2012) Structure, function and diversity of the healthy human microbiome. Nature 486(7402): 207-214.
- Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26(19): 2460-2461.
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, et al. (2013) The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res 41: D590-D596.
- (2020) R Core Team R: The R Project for Statistical Computing.
- (2020) R Studio Team, RStudio: Integrated Development for R. RStudio, PBC, Boston, USA.
- Andersen KS, Kirkegaard RH, Karst SM, Albertsen M (2018) ampvis2: An R package to analyse and visualise 16S rRNA amplicon data. bioRxiv, pp. 299537.
- Wickham H (2016) ggplot2: Elegent Graphics for Data Analysis. Springer Verlag, New York, USA.
- Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, et al. (2012) Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol 13(9): R79.
- Lopez-Siles M, Duncan SH, Garcia-Gil LJ, Martinez-Medina M (2017) Faecalibacterium prausnitzii: From microbiology to diagnostics and prognostics. ISME Journal 11(4): 841-852.
- Iljazovic A, Roy U, Gálvez EJC, Lesker TR, Zhao B, et al. (2020) Perturbation of the gut microbiome by Prevotella spp. enhances host susceptibility to mucosal inflammation. Mucosal Immunol 14: 113-124.
- Wright DP, Rosendale DI, Roberton AM (2000) Prevotella enzymes involved in mucin oligosaccharide degradation and evidence for a small operon of genes expressed during growth on mucin. FEMS Microbiol Lett 190(1): 73-79.
- Torres AG, Redford P, Welch RA, Payne SM (2001) TonB-dependent systems of uropathogenic escherichia coli: Aerobactin and heme transport and TonB are required for virulence in the mouse. Infect Immun 69(10): 6179-6185.
© 2023 Jeppe Lund Nielsen. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






