- Submissions

Full Text
Gastroenterology Medicine & Research
Cloning, Expression, and Dimer Formation Assay of GANAB cDNA in Lung Cancer Cells
Junhua Li1, Dezhi Yang1, Huricha2, Zhang2, Sachura Baoyin2, Qinglan Bao2, Du leng2, Tegexibaiyin wang1 and Wu Ji1*
1PLA Research Institute of General Surgery, Jinling Hospital, the First School of Clinical Medicine, Southern Medical University, China
2Pharmacy laboratory, Inner Mongolia International Mongolian hospital, China
*Corresponding author: Wu Ji, PLA Research Institute of General Surgery, Jinling Hospital, the First School of Clinical Medicine, Southern Medical University, Nanjing 210002, Jiangsu, China
Submission:October 11, 2021;Published: November 15, 2021

ISSN 2637-7632Volume6 Issue2
Abstract
Background: Glucosidase II is an endoplasmic reticulum heterodimer enzyme, a neutral glucosidase, involved in the trading and folding of newly synthesized glycoproteins in the endoplasmic reticulum. The destruction of GANAB leads to the accumulation of misfolded glycoproteins and the induction of unfolded protein responses. GANAB is also a key regulator of glycosylation. The encoded GANAB protein has an increased expression level in lung tumor tissues, but its function in lung cancer is not yet clear. The cloned sequence is the mutant gene of GANAB gene (NM_198334.3) in the Genbank database, and its role in lung cancer.
Method: In this study, the GANAB gene was obtained by homologous cloning, and the correct gene sequence was verified by sequencing by bioinformatics analysis; and the cDNA of the GANAB gene was cloned by RT-PCR. Aiming at high expression level, use the T base of pMD19-T Vector and the A base of the RT-PCR amplified cDNA to match the GANAB cDNA, and use the pcDNA3.1 plasmid to construct a high expression GANAB plasmid, and then use the sensory cell plasmid to purify. Western blot analysis was used to confirm the expression of the protein. Result: The cDNA fragments of 1.8kbp and 1.0kbp were isolated by RT-PCR experiment. Use pMD19-T Vector to connect the GANAB cDNA fragment to construct a 5.5kbp cDNA sequence and use the pcDNA3.1 plasmid to construct a GANAB plasmid to obtain an 8.2kbp cDNA sequence. Western blot analysis confirmed the expression of the protein. Conclusion: In this study, we cloned the GANAB gene in A549 cells and successfully introduced the forced expression into A549 cells. The cloning and protein expression of GANAB gene are of great significance to the study of the key effects of lung cancer adhesion, invasion and metastasis.Introduction
Neutral alpha-glucosidase AB (GANAB), an enzyme encoded by the GANAB gene in the
human body. The alpha subunit glucosidase II encoded by this gene is a member of the 31
group of glycosyl hydrolases. Cytogenetic location of GANAB gene: 11q12.3, the heterodimeric
GANAB separates glucose residues from immature glycoproteins in the endoplasmic
reticulum and plays a role in protein folding and quality control. The encoded GANAB protein
has an increased expression level in lung tumor tissues and reacts under ultraviolet radiation,
and mutations in this gene can cause polycystic kidney and liver diseases. Related pathways
of GANAB gene include protein metabolism, transport to the Golgi apparatus, and so on. The
gene ontology related to this gene is annotated with hydrolase activity, hydrolysis of carbonyl
compounds and so on. An important homolog of this gene is glucosidase IIβ, which is encoded
by GANC. The two subunits of GANAB and GANC play an important role in the completion
of the normal function of glucosidase II. The GenBank database shows that there are 10
mutants in the GANAB gene. GANAB is related to the occurrence of lung cancer by negatively
regulating the migration and invasion of cells. This down-regulation further supports the correlation between GANAB expression and the aggressiveness and
low survival rate of lung cancer [1-5].
Since the N-glycosylation of a variety of cellular proteins may
change the invasion and metastasis ability of tumors, GANAB may
participate in the occurrence of lung cancer by regulating the
N-glycosylation of specific target proteins. Most secreted proteins
and transmembrane proteins translocate in the endoplasmic
reticulum and undergo protein folding process and quality
control in the endoplasmic reticulum. Glucosidase II is involved in
glycoprotein processing and promotes protein folding by catalyzing
the hydrolysis of glucose residues in glycans bound to proteins.
A common feature of malignant tumors is that the glycosylation
modification of cells has undergone significant changes. In the
process of tumor occurrence and development, the glycosylation
modification of certain proteins will change with the progress of
the disease. The change of glycosylation form of glycoprotein on the
surface of tumor cells plays an important role in the process of tumor
cell adhesion, invasion and metastasis. Therefore, searching for the
characteristic changes and functional research of GANAB gene can
provide important information for the early diagnosis, progress
monitoring, prognostic evaluation and the search for therapeutic
targets of lung cancer. The study of GANAB on the changes of the
characteristic sugar chain structure of lung cancer cell membrane
and its regulation mechanism, this research has important value for
the clinical diagnosis and treatment of lung cancer.
Materials and Methods
Plasmids
pcDNA3.1 Vector is a gift from Professor OKIO HINO (Juntendo University, Japan) and pMD19-T Vector is a gift from Professor Zhang Chenyu.
Cloning of GANAB specific fragments
Extraction of total RNA from A549 cells, 1. PCR amplification
of GANAB cDNA and purification and recovery: The total RNA
of A549 cells was extracted using the Trizol extraction kit from
Invitrogen. The extracted RNA was analyzed by formaldehyde
denaturing gel electrophoresis and quantitative analysis. In this
experiment, AMV reverse transcription kit was used to synthesize
cDNA from mRNA extracted from A549 cells [5-10]. Primer 6.0
software was used to design primers. The primers were designed
according to the cDNA (NM_198334.3) sequence of the GANAB
gene published in the Genbank database. Both ends of the primer
are added with restriction bases for restriction site and restriction
site respectively. The designed pair of primers are as follows: P1
fragment upstream primer: 5’-AACTCTGCAC AAGATGGCGG-3’;
Downstream primer: 5’-GTTAGCCCACCAGGCCCT-3’. The upstream
primer of the P2 fragment: 5’-AGGGCCTGGTGGGCTAAC-3’; the
downstream primer: 5’-AACTCTGCAC AAGATGGCGG-3’. The
primers were synthesized by Beijing SBS Genetech Co., Ltd.
The synthesized first-strand cDNA is used as a template, and
the designed P1 and P2 are used as primers for amplification. The
PCR amplification conditions were 94°C pre-denaturation for 1min,
94°C for 30s, 55°C for 30s, 72°C for 3min, 35 cycles. Take 5μL of PCR
product and perform 1% agarose gel electrophoresis under 100V
voltage for 40min. DNA bands of corresponding sizes are excised
from the gel, digested, purified, electrophoresed and quantitatively
analyzed. Then the P1 cDNA fragment was digested with HindIII
and NsbI enzymes. The P2 cDNA fragment was digested with NsbI
and NSacII enzymes, and then electrophoresed on a 1% agarose
gel. The corresponding size DNA bands were excised from the gel,
digested, purified, electrophoresed and quantitatively analyzed.
Connection of PCR product and pMD19-T vector: The
recovered and purified PCR products P1 and P2 were cloned
into pMD19-T vector to construct recombinant plasmids. The
constructed plasmid was introduced into the sensory cells, and
then cultured with 200μg/ml Amp cold day medium to select the
plasmid-introduced sensory cells and cultured overnight. Then the
DNA is extracted and the extracted recombinant plasmid is used as
a template and P1 and P2 are used as primers for PCR amplification
and identification. The recombinant plasmid was entrusted to
Shanghai Sangon Biotech Co., Ltd. for sequencing and identification.
pMD19-T Vector Primer: F: 5’-CGCCAGGGTTTTCCCAGTC-3’; R:
5’-GAGCGGATAACAATTTCACACAGGAAA-3’. The sequences of all
structures were confirmed by Sanger sequencing.
Construction of pcDNA3.1-GANAB DNA plasmid: Total
DNA was extracted from A549 lung cancer cells, and then GANAB
cDNA fragments were cloned and purified using GANAB’s specific
Primer. In the second step, use pMD19-T Vector to insert the cloned
GANAB cDNA fragment into the EcoR V enzyme digestion position,
and then use the sensory cell dH5α to amplify the recombinant
GANAB expression plasmid, and then perform gel electrophoresis
to extract from the gel The P1 fragment was digested with HindIII
and NsbI digestive enzymes; the P2 fragment was extracted from
the gel and then digested with NsbI and SacII digestive enzymes.
At the same time, pcDNA3.1 plasmid was digested by HindIII
and SacII. Then, insert P1 and P2 GANAB cDNA between the
Hind III and SacII digestive enzyme sites of pcDNA3.1 Vector.
Amplify the recombinant GANAB expression plasmid with DH5α
(Introin in the United States) of the constructed plasmid and
use specific primers to sequence it firmly. The primers are as
follows: F: 5’-CGGAATTCTACTCTATATACATGCTCCATCC-3’, R:
5’-CGTCTAGATCACAGACCAAGAGAGAAAGGAAAC-3’. The sequences
of all structures were confirmed by Sanger sequencing.
Verification of the protein expression of the constructed
plasmid: A549 cells are seeded in a 6-well plate at a concentration
of 106 cells/well and transfected when the cells have grown to
30%-50%. The experiment is divided into 2 groups: pcDNA3.1-
GANAB group and mock group. Prepare two sterile 1.5ml EP tubes,
add 250μl RPMI1640 medium and 5μl lipofectamin 2000 to each;
then, add 5μl pcDNA3.1-GANAB to one EP tube, and mock plasmid
to the other EP tube, and mix them well at room temperature
After standing for 5 minutes, transfer to 4°C for storage. Then discard the supernatant of the cultured cells in the 6-well plate,
add 1ml of serum-free RPMI1640 medium to each well, then add
the prepared plasmid mixture to each well, mix it, and incubate at
37℃, 5% CO2 Cultivate in the box for 4-6h. Discard the transfection
solution, add 1.5ml of RPMI1640 medium containing 10% FBS to
each well and incubate for 48 hours, wash twice with 1×PBS, add
750μl RIPA buffer to fully lyse; then use western blotting to detect
the expression of GANAB protein Condition. (Primary antibody:
Secondary antibody:)
Result
PCR amplification of human GANAB gene cDNA
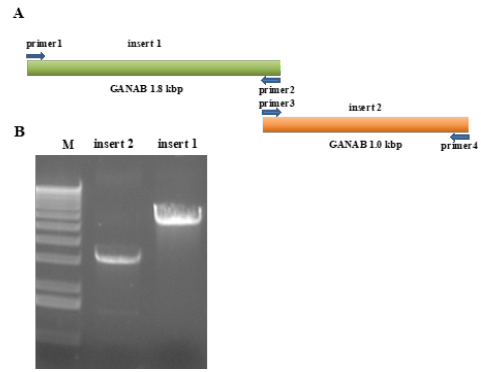
Synthesize the cDNA of human GANAB gene with the GANAB gene (NM_198334.3) sequence in Genbank as a template. In this experiment, the cDNA of the GANAB gene was divided into two fragments for cloning, namely the P1 fragment and the P2 fragment. After using 1% agarose gel electrophoresis to excise the 1800bp and 1000bp specific amplified bands, the size of our amplified GANAB cDNA is 2834bp (Figure 1 & 2).
Figure 1: Cloning of ganab gene. PcDNA3.1-GANAB expression plasmid was constructed with pmd19-t and pcDNA3.1 plasmids.
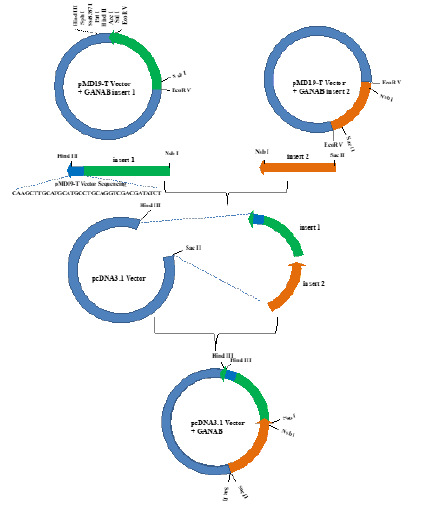
Figure 2: PCR amplification of ganab cDNA sequence.
A. The ganab cDNA sequence is divided into insert 1 and insert 2.
B. Results of PCR amplification of ganab cDNA sequence insert 1 and insert 2.
Cloning of pMD19-T GANAB plasmid
The amplified GANAB cDNA P1 and P2 fragments were cloned using pMD19-T plasmid. Sequencing verification using T vector specific primers. After the ligation is successful, the plasmid is digested with EcoR V enzyme, and after electrophoresis on a 1% agarose gel, there should be a single DNA band around 3.8kbp and 2.8kbp. Then use a DNA sequencer to sequence the results consistent with the involved sequence. It is proved that the GANAB gene cDNA P1 and P2 fragments are successfully connected with the T vector (Figure 3 & 4).
Figure 3: M: Maeker X.
1. GANAB cDNA insert 2.8kbp.
2. pMD19-T Vector+GANAB 5.5kbp.
3. pcDNA3.1 GANAB plasmid 8.2kbp.
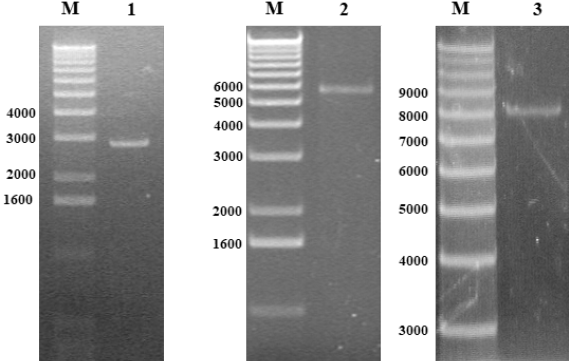
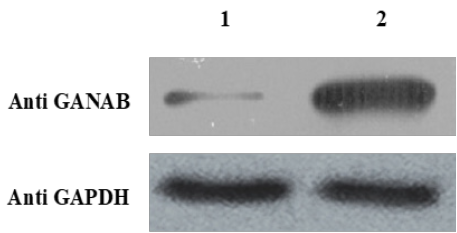
Figure 4: Expression of ganab plasmid.
1. Expression of pcDNA3.1 mock plasmid.
2. Expression of pcdna3.1-ganab plasmid.
Construction of GANAB expression vector
Total RNA was extracted from A549 cells, and specific primers were used to amplify the P1 and P2 fragments of GANAB cDNA, and then the P1 and P2 fragments were inserted into the EcoR V site of the pMD19-T plasmid. The GANAB coding sequence was cut out and cloned into the Hind III/Sac II site of pcDNA3.1. In order to establish the correct reading frame, the resulting plasmid was digested with Sac II, and then Hind III/NsbI digested GANAB cDNA P1 and NsbI/ Sac II digested GANAB cDNA P2 fragments were added to reconnect [10-17]. This plasmid is called pcDNA3.1-GANAB. The constructed plasmid pcDNA3.1-GANAB was digested and cut with Hind III enzyme, and a 8.2kbp fragment was observed by 1% agarose gel electrophoresis. Then use a DNA sequencer to sequence the results consistent with the involved sequence. Prove that the construction of pcDNA3.1-GANAB was successful (Figure 3).
Construction of the protein expression of the plasmid
A549 cells were transfected with pcDNA3.1-GANAB plasmid and mock plasmid. Incubate in a 37°C, 5% CO2 incubator for 4-6 hours. Discard the transfection solution, add 1.5ml of RPMI1640 medium containing 10% FBS to each well and incubate for 48 hours, wash twice with 1×PBS, add 750μl RIPA buffer to fully lyse; then use western blotting to detect the expression of GANAB protein Condition. (Primary antibody: Secondary antibody:)
Discussion
GANAB, also known as glucosidase II-α subunit, is a neutral
glucosidase that participates in the trading and folding of newly
synthesized glycoproteins in the endoplasmic reticulum. The
destruction of GANAB leads to the accumulation of misfolded
glycoproteins and the induction of unfolded protein responses.
GANAB is also a key regulator of glycosylation. The absence
of GANAB caused the inhibition of the biosynthesis of new
N-glycosylated proteins on the cell surface. According to reports,
the lack of glycosidase II is related to polycystic liver disease. In
polycystic liver disease, hepatic cystine cannot bind to GANAB
during carbohydrate processing, leading to changes in cell
proliferation and differentiation. Except for polycystic liver disease,
GANAB has not been reported to be related to other diseases. In
this study, it was found that the encoded GANAB protein has
increased expression levels in lung cancer tissues and cells. GANAB
is related to the occurrence of lung cancer by negatively regulating
the migration and invasion of cells. This down-regulation further
supports the correlation between GANAB expression and the
aggressiveness and low survival rate of lung cancer. Since the
N-glycosylation of a variety of cellular proteins may change the
invasion and metastasis ability of tumors, GANAB may participate
in the occurrence of lung cancer by regulating the N-glycosylation
of specific target proteins.
The GenBank database shows that there are 10 mutants in the
GANAB gene. The rapid increase in the number of cloned genes has
led to an increase in the ability to recognize sequence homology
between different genes, whether within the same species
(homologs) or between species (homologs). Easier identification
and cloning of gene family member genes in electrophoresis
provides the potential to greatly facilitate their physical cloning.
Our research report cloned the previously described human
neutral a-glucosidase AB gene (GANAB), which is responsible for
the catabolism and regulation of carbohydrate chain degradation.
These enzymes are mainly identified as members of Glycoside
Hydrolase (GH) family 13 and 31. The GH13 family contains a
variety of glucoside processing enzymes. The combination of human
gene research and cell analysis shows that this study obtained
the GANAB gene through homologous cloning and performed
bioinformatics analysis on the correct gene sequence verified by
sequencing; and through RT-PCR, Analyzed the mRNA expression
pattern of GANAB gene, and verified the protein expression of
this gene by plasmid overexpression experiment and western blot
experiment. It is expected to lay the foundation for further research
on the molecular mechanism and biological function of the gene
expression regulation.
References
- Porath B, Gainullin VG, Cornec-Le Gall E, Dillinger EK, et al. (2016) Mutations in GANAB, encoding the glucosidase IIα subunit, cause autosomal-dominant polycystic kidney and liver disease. Am J Hum Genet 98(6): 1193-1207.
- van de Laarschot LFM, Te Morsche RHM, Hoischen A, Venselaar H, Roelofs HM, et. al. (2020) Novel GANAB variants associated with polycystic liver disease. Orphanet J Rare Dis 15(1): 302.
- Geng G, Xiao Y, Zhang Y, Shen W, Liu J, et al. (2020) Ganab haploinsufficiency does not cause polycystic kidney disease or polycystic liver disease in mice. Biomed Res Int, pp. 7469428.
- Gabriško M (2020) The in silico characterization of neutral alpha-glucosidase C (GANC) and its evolution from GANAB. Gene 726: 144192.
- Waldrop E, Al-Obaide MAI, Vasylyeva TL (2019) GANAB and PKD1 variations in a 12 years old female patient with early onset of autosomal dominant polycystic kidney disease. Front Genet 10: 44.
- Wilson EM, Choi J, Torres VE, Somlo S, Besse W (2020) Large deletions in GANAB and SEC63 explain 2 cases of polycystic kidney and liver disease. Kidney Int Rep 5(5): 727-731.
- Besse W, Choi J, Ahram D, Mane S, Sanna-Cherchi S, et al. (2018) A noncoding variant in GANAB explains isolated polycystic liver disease (PCLD) in a large family. Hum Mutat 39(3): 378-382.
- Qiao Q, Bouwman FG, van Baak MA, Roumans NJT, Vink RG, et al. (2019) Adipocyte abundances of CES1, CRYAB, ENO1 and GANAB are modified in-vitro by glucose restriction and are associated with cellular remodeling during weight regain. Adipocyte 8(1): 190-200.
- Levy-Ontman O, Fisher M, Shotland Y, Tekoah Y, Malis Arad S (2015) Insight into glucosidase II from the red marine microalga Porphyridium sp. (Rhodophyta). J Phycol 51(6): 1075-87.
- Rauscher B, Heigwer F, Henkel L, Hielscher T, Voloshanenko O, et al. (2018) Toward an integrated map of genetic interactions in cancer cells. Mol Syst Biol 14(2): e7656.
- Al-Hamed MH, Alsahan N, Rice SJ, Edwards N, Nooreddeen E, et al. (2019) Bialleleic PKD1 mutations underlie early-onset autosomal dominant polycystic kidney disease in Saudi Arabian families. Pediatr Nephrol 34(9): 1615-1623.
- Cordido A, Besada-Cerecedo L, García-González MA (2017) The Genetic and cellular basis of autosomal dominant polycystic kidney disease-a primer for clinicians. Front Pediatr 5: 279.
- Qin Y, Zhao L, Wang X, Tong D, Hoover C, et al. (2017) MeCP2 regulated glycogenes contribute to proliferation and apoptosis of gastric cancer cells. Glycobiology 27(4): 306-317.
- Chiu CC, Lin CY, Lee LY, Chen YJ, Lu YC, et al. (2011) Molecular chaperones as a common set of proteins that regulate the invasion phenotype of head and neck cancer. Clin Cancer Res 17(14): 4629-4641.
- Wills ES, Te Morsche RHM, van Reeuwijk J, Horn N, Geomini I, et al. (2017) Liver cyst gene knockout in cholangiocytes inhibits cilium formation and Wnt signaling. Hum Mol Genet 26(21): 4190-4202.
- Cornec-Le Gall E, Chebib FT, Madsen CD, Senum SR, Heyer CM, et al. (2018) The value of genetic testing in polycystic kidney diseases illustrated by a family with PKD2 and COL4A1 Am J Kidney Dis 72(2): 302-308.
- Wang J, Yang H, Guo R, Sang X, Mao Y (2021) Association of a novel PKHD1 mutation in a family with autosomal dominant polycystic liver disease. Ann Transl Med 9(2): 120.
© 2021 Wu Ji. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






