- Submissions

Full Text
Gastroenterology Medicine & Research
Resistance To Beta-Lactamics in Gastroenteric Processes in the Brazilian Amazon
Daniela Cristiane da Cruz Rocha, Érica Kássia Sousa Vidal, Débora de Castro Costa, Karina Lúcia Silva da Silva and Anderson Nonato do Rosário Marinho*
Evandro Chagas Institute, Laboratory of Bacterial Enteroinfections, Brazil
*Corresponding author:Anderson Nonato do Rosário Marinho, Evandro Chagas Institute, Laboratory of Bacterial Enteroinfections, Brazil
Submission: March 01, 2021;Published: March 23, 2021

ISSN 2637-7632Volume5 Issue5
Abstract
In recent years there has been a rapid increase in bacterial resistance in Brazil and worldwide. Among the mechanisms described, Extended-Spectrum ß-Lactamases (ESBL) are of great importance. The continuous assessment of bacterial resistance have fundamental importance in preventing the occurrence of outbreaks and the spread of resistance, especially in the Amazon due to the relative isolation of cities and difficulties in epidemiological surveillance in the region. In the present work we approach the use of phenotypic and genotypic characterization in Enterobacterial isolates by determining the resistance profile to β-lactam antimicrobials by antibiogram and the search for blaCTX-M, blaTEM, blaSHV genes associated with β-lactam resistance. Thus, 88 samples from outpatients from 14 regions in northern Brazil, belonging to the Brazilian Amazon, were analyzed from 2012 to 2014. Of these, Salmonella Typhi (57/64.77%), Salmonella spp. (26/29.54%), Shigella flexneri (2/2.27%), Shigella dysenterie (1/1.14%), Shigella sonnei (1/1.14%) and diarrhogenic Escherichia coli (1/1.14%). As for the resistance profile, only Escherichia coli showed resistance to first, second, third and fourth generation penicillins and cephalosporins and the aztreonam monobactamide, the other isolates did not show resistance profile according to the Clinical and Laboratory Standards (CLSI) standards. The evaluation of the blaCTX-M, blaTEM and blaSHV genes showed positive results only for the blaCTX-M gene in the Escherichia coli sample, with no amplifications being observed in the other analyzed samples. Although resistance was observed in only one sample, the data suggest the circulation of β-lactam resistance genes in Enterobacteria and, therefore, we suggest that continuous resistance evaluation can prevent the spread of β-lactamase-producing Enterobacteria, evidencing the importance of continuous monitoring and efficient strategies to reduce the spread of these agents by preventing outbreaks by resistant strains.
Keywords: β-lactamases; ESBLs; Antimicrobial resistance; Enterobacteria, PCR
Introduction
In recent years there has been a rapid increase in bacterial resistance in Brazil and worldwide, the occurrence of this phenomenon in regions such as the Brazilian Amazon is still poorly known, but they are already observed in populations in northern South America in hospital cases and in localized outbreaks [1,2]. The production of Extended Spectrum Β-Lactamase (ESBLs) is an important mechanism of resistance in Enterobacteria. The emergence and spread of ESBL among family members has been described worldwide as a point of clinical urgency due to the high incidence of these isolates in infections related to health care, such as urinary and intestinal infections, pneumonia, septicemia, meningitis, among others [3,4]. The main genera producing ESBLs, among the Enterobacteria, are Escherichia coli and Klebisiella pneumoniae [5]. Currently, increasing resistance to antimicrobials is a highly complex problem for global health that is often associated with the widespread and sometimes indiscriminate use of antimicrobials that work in the selection and dissemination of resistance [6-8]. Resistance to the vast majority of beta-lactams, including carbapenemics in ESBL-producing and KPC-type bacteria, is often concomitantly associated with resistance to other classes of antimicrobials, such as aminoglycosides and fluorquinolones. Because of this, the rapid identification of strains that produce these enzymes is of fundamental importance in the selection of the appropriate antimicrobial for the treatment [9-11].
The need for speedy identification of antimicrobial therapy is due to the fact that bacteria have a great capacity to conserve and transmit plasmids, as well as other mobile elements that contain resistance genes, such as beta-lactamase genes, thus facilitating the spread of Anti Microbial Resistance (AMR). The presence of these resistance mechanisms has a direct interference in the treatment of the patient, reducing the therapeutic alternatives available, arising the need to resort to the latest generation of antimicrobials, which increases the selective pressure and can cause the appearance of new types of resistance, restricting increasingly possible treatment options [12-16].
Methods
88 pathogenic strains isolated from species belonging to the Enterobacteriaceae family were analyzed, collected from 2012 to 2014 (2012-27 samples; 2013-19 samples and 2014-42 samples) from clinical cases treated in the Brazilian Amazon from 14 municipalities outpatients infected with pathogenic Enterobacteria. CAAE Ethics Committee: 52530916.0.0000.0019 Instituto Evandro Chagas (CEP/IEC/SVS/MS). The susceptibility to antimicrobial agents was assessed by the disc diffusion method according to the recommendations of the Clinical and Laboratory Standards Institute- (CLSI, 2012) [17] and by the automated system Vitek 2 (BioMèrieux) following the manufacturer's recommendations. As controls for the susceptibility tests to antimicrobials, the strains American Type Culture Collection (ATCC), K. pneumoniae ATCC 700603 producer of ESBL and E. coli ATCC 25922 were used. For the extraction of bacterial DNA, the DNA IQ kit (Promega) was used, following the manufacturer's recommendations. For the amplification of the blaCTX-M, blaTEM, blaSHV genes, the primers described by Schmitt et al. [18], Nasehi et al. [19] and Edelstein et al. [20] were used. Each amplification reaction had a final volume 25μL, containing 20ng of DNA, 10Mm Tris-HCl, pH 8.5, 50mMKCl, 1.5/μM MgCl2, 1.25mM of each dNTP, 1.25mM of each primer and 0.5 unit of Taq DNA polymerase Platinum (Invitrogen). The amplifications were performed in a Vereti ™ 96-Well Thermal Cycler thermocycler (Applied Biosystems-US). After amplification, the samples were applied to a 2% agarose gel, observed in an ultraviolet transluminator and recorded in Bioimaging Systems (UPV, USA) detection systems, the molecular weight of the amplified fragments was measured using a 1Kb molecular weight marker, along with the positive and negative controls.
Results
It was observed that the occurrence of cases had a higher percentage distribution for males (57.35%) and the prevalent frequence among adults (67.04%) (Table 1). Among the isolates, the genus Salmonella was predominantly observed in 83(94.32%) of the 88 isolates belonging to this genus. Of these 57(64.77%) were identified as Salmonella Typhi and 26(29.54%) as Salmonella spp (Table 2). The results of the antimicrobial susceptibility profile showed that Escherichia coli was the only microorganism that showed resistance to penicillins and first, second, third and fourth generation cephalosporins and the monobactamide aztreonam. While Salmonella Typhi, Salmonella spp, Shigella flexneri, Shigella dysenterie and Shigella sonnei, did not present a resistance profile according to the CLSI (2012) [17] (Table 3). After PCR amplification to assess the presence of the ESBLs encoding genes blaCTX-M, blaTEM and blaSHV, amplifications were observed exclusively for the CTX-M gene and only in Escherichia coli (Figure 1), with no amplifications for the blaTEM and blaSHV investigated. All other samples analyzed did not show amplifications for the genes encoding ESBLs (blaCTX-M, blaTEM and blaSHV) analyzed.
Table 1:Age/gender distribution of patients infected with Enterobacteria between January 2012 and December 2014.

N.I= Not Informed
Table 2: Frequency of isolated microorganisms belonging to the Enterobacteriaceae family from 2012 to 2014.

Table 3: Profile of susceptibility to antimicrobials among samples belonging to the Enterobacteriaceae family.
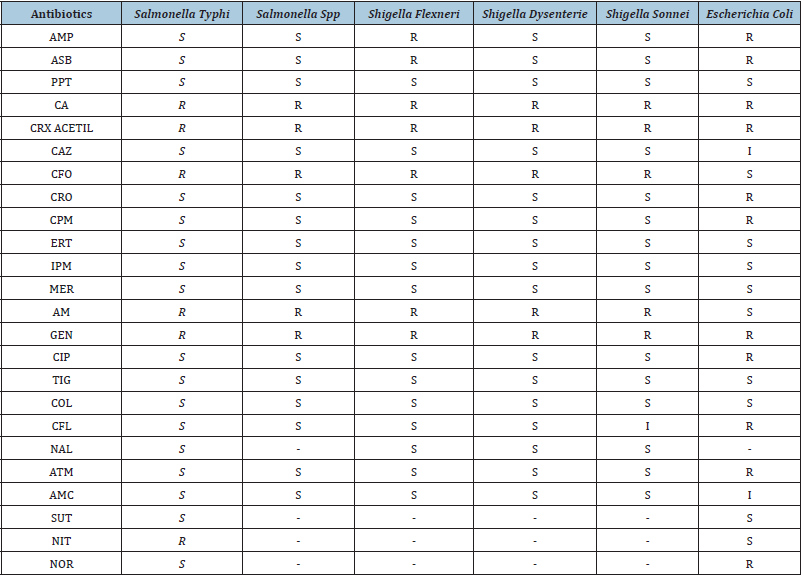
S=Sensitive; R=Resistant; AMP=Ampicillin; ASB=Ampicillin + Sulbactam; PPT=Piperacillin + Tazobactam; CRX=Cefuroxime; CA=Cefuroxime axetil; CAZ=Ceftazidime; CFO=Cefoxitin; CRO=Ceftriaxone; CPM=Cefepime; ERT=Ertapenem; IPM=Imipenem; MER=Meropenem; AM=Amikacin; GEN=Gentamycin; CIP=Ciprofloxacin; TIG=Tigecycline; COL=Colistin; CFL=Cephalothin; NAL=Nalidixic acid; ATM=Aztreonam; AMC=Amoxacillin + Clavulanic Acid; SUT=Sulfamethoxazole + Trimetroprim; NIT=Nitrofurantoin; NOR=Norfloxacin.
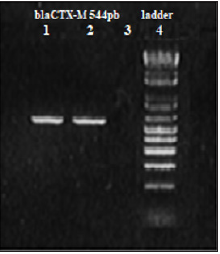
Figure 1:-2% agarose gel, blaCTX-M amplification in Escherichia coli.
A. Line 1: Positive control.
B. Line 2: blaCTX-M in E. coli.
C. Line 3: negative control.
D. Line 4: DNA Ladder 1Kb.
Discussion
Diarrheal cases are frequent in the Brazilian Amazon, especially in rainy periods [21], monitoring these cases is crucial to determine mitigation measures in the populations in order to prevent outbreaks [6]. Although samples of Salmonella Typhi, Salmonella spp, Shigella flexneri, Shigella dysenterie, Shigella sonnei have been shown to be sensitive to first and second generation cephalosporins and aminoglycosides, according to the CLSI (2012) [17]. For periodic analysis the phenotypic and molecular profile of antimicrobial resistance genes are crucial for the early detection of the horizontal transmission of the genetic components encoding ESBLs. [22-24]. Similar to the evidence in the early 2000s, where a study carried out on Shigella strains demonstrated resistance to tetracycline (93.4%) followed by chloramphenicol (63.9%), trimethoprim/sulfamethoxazole (63.1) and ampicillin (43.4%) [25] that previously identified allowed a quick and concrete action in the population. Diarrheal cases caused by diarrheal E. coli are often associated with children under 5 years old [26-28] and are a serious health problem in the Amazon. The observation of a sample of Escherichia coli producing ESBL by the blaCTX-M gene, with resistance to first to fourth generation cephalosporins, penicillin, aminoglycosides, fluoroquinolones, quinolones and aztreonam demonstrates the circulation of this gene in samples in the studied population. This shows that the detected E. coli has high levels of resistance to antibiotics currently prescribed, which leads to the therapeutic choice of carbapenem antibiotics. In a study involving 354 clinical isolates of E. coli, from five outpatient and hospital units, from October 2002 to May 2003, in Rio de Janeiro, 8 isolates producing ESBL were found, that is, in 2.2 % of those [29]. Nogueira et al. [30] studied 498 isolates from patients at a university hospital in Curitiba, from 2003 to 2004, and determined that 7.2% were E. coli producing ESBL, that is, 2.2% of these. In the same sense, Lago et al. [3] in their study of 838 bacterial isolates, from patients hospitalized in Passo Fundo, RS, from July to December 2007, 96 E. coli were identified and 11.4% of these were in our studies, in the molecular detection of the genes encoding the enzymes ESBL, the PCR technique revealed a gene of the researched ESBLs of the CTX-M type. Enzymes belonging to the CTX-M family have been predominant in South America, as well as in Spain and Eastern Europe [31]. The data show an increase in the percentage of isolation of ESBL-producing E. coli in Brazil, and the same can occur in the Brazilian Amazon, but the lack of studies in populations and the great distances associated with the isolation of cities makes surveillance difficult or unviable, however, monitoring as proposed in the present study can ensure a minimum of surveillance to determine the need for more concrete actions in the care of outbreaks as well as sporadic cases.
Conclusion
The results obtained through this study demonstrated the permanence and the spread of Enterobacteria that cause gastroenteritis, showing the importance of a continuous evaluation and efficient strategies to reduce the spread of these agents. The production of extended-spectrum beta-lactamases was evidenced, even if in a low proportion, this data is of concern, since in the clinic the production of ESBLs limits the effectiveness of beta-lactam antibiotics including extended-spectrum cephalosporins.
References
- Baizet C, Ouar-Epelboin S, Walter G, Mosnier E, Moreau B, et al. (2019) Decreased antibiotic susceptibility of Enterobacteriaceae causing community-acquired urinary tract infections in French Amazonia. Med Mal Infect 49(1): 63-68.
- Martínez-Puchol S, Riveros M, Ruidias K, Granda A, Ruiz-Roldán L, et al. (2021) Dissemination of a multidrug resistant CTX-M-65 producer Salmonella entérica serovar infantis clone between marketed chicken meat and children. International Journal of Food Microbiology 344: 109109
- Lago A, Fuentefria SR, Fuentefria DB (2010) ESBL-producing Enterobacteria in Passo Fundo, state of Rio Grande do Sul, Brazil. Rev Soc Bras Med Trop 43 (4): 430-434.
- Silva KC, Lincopan N (2012) Epidemiology of extended-spectrum beta-lactamases in Brazil: Clinical impact and implications for agribusiness. Journal Bras Patol Med Lab 48(2): 91-99.
- Janda JM, Abbott SL (2005) The Enterobacteria. (2nd edn), American society for microbiology, Washington, USA, pp. 411.
- Bulteel AJB, Larson EL, Getahun H (2020) Identifying global research gaps to mitigate antimicrobial resistance: A scoping review. Am J Infect Control 28(20): 31033-31036.
- Morrison L, Zembower TR (2020) Antimicrobial resistance. Gastrointest Endosc Clin N Am 30(4): 619-635.
- Júnior MAS, Ferreira ES, Conceição GC (2004) Extended Spectrum Betalactamases (ESBL): An important mechanism of bacterial resistance and its detection in the clinical laboratory. Newslab 63: 152-172.
- Wuerz TC, Kassim SS, Atkins KE (2020) Acquisition of extended-spectrum beta-lactamase-producing Enterobacteriaceae (ESBL-PE) carriage after exposure to systemic antimicrobials during travel: Systematic review and meta-analysis. Travel Med Infect Dis 37: 101823.
- Villegas MV, Blanco MG, Osornio JS, Rossi F (2011) Increasing prevalence of extended-spectrum-betalactamase among gram-negative bacilli in latin America-2008 update from the Study for Monitoring Antimicrobial Resistance Trends (SMART). Braz J Infect Dis 15(1): 34-39.
- Blatt JM (2000) Mechanism of resistance and detection of broad-spectrum beta-lactamases. Newslab 7(1): 59-63.
- Edwards T, Williams C, Teethaisong Y, Sealey J, Sasaki S, et al. (2020) A highly multiplexed melt-curve assay for detecting the most prevalent carbapenemase, ESBL, and AmpC genes. Diagn Microbiol Infect Dis 97(4):115076.
- Motamedifara M, Mohebib S, Hadadib M, Amirzadegan H (2020) The prevalence of ESBL and AmpC β-lactamases in uropathogenic isolates of Escherichia coli in a tertiary care hospital in Southwest Iran. Gene Reports 20: 100747.
- Hansen SK, Kaya H, Roer L, Hansen F, Skovgaard S, et al. (2020) Molecular characterization of danish ESBL/AmpC-producing Klebsiella pneumoniae from bloodstream infections, 2018. J Glob Antimicrob Resist 22: 562-567.
- Chiang CY, Uzoma I, Moore RT, Gilbert M, Duplantier AJ, et al. (2018) Mitigating the impact of antibacterial drug resistance through host-directed therapies: Current progress, outlook, and challenges. mBio 9(1): e01932-17.
- Macgowan AP (2008) Clinical implications of antimicrobial resistance for therapy. J Antimicrob Chemother 62(2): 105-114.
- Clinical and Laboratory Standards Institute (CLSI) document for antimicrobial susceptibility testing. Informational supplement document M100 S15. CLSI- Pennsylvania, USA.
- Schmitt J, Jacobs E, Schmidt H (2007) Molecular characterization of extended-spectrum beta-lactamases in Enterobacteriaceae from patients of two hospitals in Saxony, Germany. J Clin Microbiol 56(2): 241-249.
- Nasehi L, Shahcheraghi F, Nikbin VS, Nematzadeh S (2010) PER, CTX-M, TEM and SHV beta-lactamases in clinical isolates of Klebsiella pneumoniae isolated from Tehran, Iran. Iran J Basic Med Sci 13(3): 111-118.
- Edelstein M, Pimkin M, Palagin I, Edelstein I, Stratchounski L (2003) Prevalence and molecular epidemiology of CTX-M extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in Russian hospitals. Antimicrob Agents Chemother 47(12): 3724-3732.
- Rocha DCC, Marinho ANR, Reis MSO, Borges IR, Ramos FLP, et al. (2014) Epidemiological profile and molecular characterization of Salmonella Typhi isolated in the state of Pará, Brazil. Pan-Amazonian Journal of Health 5 (4): 53-62.
- Vaidya VK (2011) Horizontal transfer of antimicrobial resistance by extended-spectrum β lactamase-producing Enterobacteriaceae. J Lab Physicians 3(1): 37-42.
- Doi Y, Adams-Haduch JM, Peleg AY, D'Agata EM (2012) The role of horizontal gene transfer in the dissemination of extended-spectrum beta-lactamase-producing Escherichia Coli and Klebsiella pneumoniae isolates in an endemic setting. Diagn Microbiol Infect Dis 74(1): 34-38.
- Daniel S, Goldlust K, Quebre V, Shen M, Lesterlin C, et al. (2020) Vertical and horizontal transmission of Esbl Plasmid from Escherichia Coli O104:H4. Genes (Basel) 11(10): 1207.
- Bastos FC, Loureiro ECB (2010) Characterization of antimicrobial resistance in samples of Shigella Isolated in Belém, state of Pará, Brazil (1990-2000). Pan-Amaz Saúde Magazine 1 (4): 71-74.
- Orlandi PP, Magalhães GF, Matos NB, Silva T, Penatti M, et al. (2006) Etiology of diarrheal infections in children of porto velho (rondônia, western amazon region, Brazil). Braz J Med Biol Res 39(4): 507-517.
- Orlandi PP, Silva T, Magalhães GF, Alves F, de Almeida Cunha RP, et al. (2001) Enteropathogens associated with diarrheal disease in infants of poor urban areas of porto velho, Rondônia: A preliminary study. Mem Inst Oswaldo Cruz 96(5): 621-625.
- Fontoura VM, Graepp-Fontoura I, Santos FS, Santos Neto M, Tavares HSA, et al. (2018) Socio-environmental factors and diarrheal diseases in under five-year old children in the state of Tocantins, Brazil. PLoS One 13(5): e0196702.
- Del Peloso PF, Barros MFL, Santos FA (2010) Sepse por serrtia marcescens KPC. J Bras Patol Med Lab 46(5): 365-367.
- Nogueira KS, Higuti IH, Nascimento AJ, Terasawa LB, Oliveira S, et al. (2006) Occurrence of extended-spectrum beta-lactamases in Enterobacteriaceae isolated from hospitalized patients in Curitiba, southern Brazil. Braz J Infect Dis 10(6): 390-395.
- Bonelli RR, Moreira BM, Picão RC (2014) Antimicrobial resistance among Enterobacteriaceae in South America: History, current dissemination status and associated socioeconomic factors. Drug Resist Updat 17(1-2): 24-36.
© 2021 Anderson Nonato do Rosário M. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






