- Submissions

Full Text
Gastroenterology Medicine & Research
Complications of Gastroenteritis By Typhoid Fever in Amazonia: Clinical Cases Genetic Evaluation from Intestinal Drilling, Pneumonia and Cholestatic Hepatitis
Daniela Cristiane da Cruz Rocha, Karina Lúcia Silva da Silva and Anderson Nonato do Rosário Marinho*
Evandro Chagas Institute, Laboratory of Bacterial Enteroinfections, Brazil
*Corresponding author: Anderson Nonato do Rosário Marinho, Evandro Chagas Institute, Laboratory of Bacterial Enteroinfections, Brazil
Submission: September 02, 2020;Published: October 06, 2020

ISSN 2637-7632Volume5 Issue2
Abstract
Typhoid fever is a systemic bacterial disease caused by Salmonella enterica serotype Typhi, manifested by prolonged fever accompanied by intestinal disorders, which in some cases may progress to intestinal perforation, and can also cause pneumonia and cholestatic hepatitis. The virulence of Salmonella Typhi is highly complex, involving the expression of numerous genes, which encode toxins, adhesins, invasions or other virulence factors. In the present work, the bacterial isolate from three clinical cases of typhoid fever complications (intestinal perforation, pneumonia and cholestatic hepatitis) were tested for six virulence genes (invA, viAB, prt, tcf, tyv and H-d). The samples showed the same results in the PCR identifying the agent as Salmonella Typhi, without the presence of other non-Typhi Salmonella, being observed the existence of high genetic similarity between the samples of the analyzed clinical cases.
Keywords: Salmonella Typhi; Virulence genes; Polymerase chain reaction (PCR)
Introduction
Typhoid fever is a systemic disease caused by Salmonella enterica serotype Typhi, characterized by high fever accompanied by intestinal disorders, whose transmission occurs through the consumption of contaminated water and food [1,2]. The method used for the diagnosis is culture, which can result in false negatives due to previous use of antibiotics. Failure to diagnose the outcome makes therapeutic conduct unfeasible, constituting an aggravating factor in the clinic than some cases. One way around this problem is to use assays based on the amplification of nucleic acids [3,4]. Typhoid fever remains one of the most important infectious diseases in the world, which has seen little decrease in mortality rates since the 1990s [5]. Countries in East and Southeast Asia, Africa, the Caribbean, Central and South America currently suffer from this public health problem [6]. The World Health Organization (WHO) estimates that there are 11 to 20 million new cases in the world and approximately 128,000 to 161,000 deaths each year [7]. In Brazil, between 2001 and 2019, 1,745 cases of typhoid fever were confirmed, the Brazilian Amazon, although it represents 59% of the Brazilian territory, but with the lowest population density, 12.83% of the total population, reported the largest number of cases at all parents with 1,491 (85.44% of the national total) [8]. Thus, the Evandro Chagas Institute (IEC) invests in the program aimed at the surveillance of gastroenteric diseases and their complications with a focus on laboratory diagnosis of patients with suspected typhoid fever from all over the Brazilian Amazon [9].
Methods
Three Salmonella Typhi bacterial isolates from blood culture and coproculture of patients with intestinal perforation, pneumonia and cholestatic hepatitis were evaluated, available at the Bacteriology and Mycology Section of the Evandro Chagas Institute (IEC). To identify the Salmonella species and its genetic profile, six virulence genes invA, viAB, prt, tcf, tyv and H-d were analyzed [3,10,11]. The DNA of the bacterial isolates was extracted using the boiling and freezing technique [12]. Each set of primers (1.25mM) was incubated in an amplification reaction with a final volume of 25μL, containing 20ng of DNA, 10Mm Tris-HCl, pH 8.5, 50mMKCl, 1.5/μMMgCl2, 1.25mM of each dNTP and 0.5 units of Taq DNA polymerase Platinum (Invitrogen), incubated in an automatic gradient thermocycler model Vereti™ 96-Well Thermal Cycler (Applied Biosystems-US), with program: four minutes at 95ºC, thirty-five one minute cycles at 95ºC, one minute at 60ºC and one minute at 72ºC, final cycle of seven minutes at 72ºC. The product was visualized on an agarose gel (2%) using a 1Kb molecular weight marker, together with the positive control (strain S57) and negatives.
Results
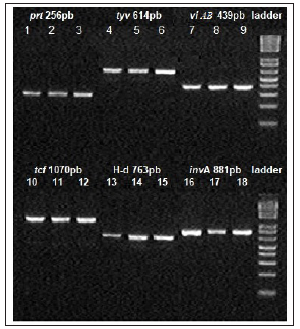
As result, it was observed that in the three clinical cases analyzed, all the investigated genes (invA, viAB, prt, tcf, tyv and H-d.) were amplified, characterizing the presence of Salmonella Typhi, with no other species or serogroup of Salmonella. There were no variations in the molecular weight of the amplicons, which demonstrated genetic similarity between all samples surveyed (Figure 1). When comparing the three clinical cases, there was also no variation between samples, with all of them showing the same amplification pattern in relation to the studied genes.
Figure 1: Electrophoresis in 2% agarose gel for visualization of the amplification product using the primers prt, tyv, viAB, tcf, H-d and invA: 1, 4, 7, 10, 13 and 16 - intestinal drilling; 2, 5, 8, 11, 14 and 17 - pneumonia; 3, 6, 9, 12, 15 and 18 - cholestatic hepatites, Ladder 1Kb. Source: IEC Collection.
Discussion
Complications of typhoid fever are rare in immunocompetent patients, being more frequent in patients with immunodeficiency syndromes, immunosuppressed transplant recipients or even in children [13-15]. The patient with intestinal perforation analyzed in the present study was seen at the Evandro Chagas Institute and diagnosed with Salmonella Typhi by the classic methods of coproculture, however the evaluation of the presence of the invA, viAB, prt, tcf, tyv and H-d genes was necessary due to the existence of other endemic agents that cause gastroenteritis in the region and the existence of 2500 Salmonella serotypes and there are no studies on the distribution of these serotypes in the Brazilian Amazon. However, the results showed that there was no other agent involved and confirmed Salmonella Typhi as the causative agent. The data for virulence genes are important since the pathogenesis of the disease is related not only to the infectious bacterial load, estimated at 106 to 109 ingested bacteria, but also to the virulence of the strain and the host’s immune response, which can facilitate or hinder the evolution of the disease to more severe cases such as intestinal perforation [16]. Thus, often associated with gastrointestinal disorders, Salmonella can cause pneumonia and cholestatic hepatitis in addition to intestinal perforation. Although historically associated with immunodeficiency [13,14], pulmonary cases of Salmonellas are also described in immunocompetent patients, who can progress to septic shock and multiple organ failure. In these cases, pneumonia can be caused only by Salmonella or in association with other bacteria or even viruses. However, in the existing studies the Salmonellas found are often Salmonella Enteritidis, Salmonella enterica serotype Choleraesuis, with rare cases associated with Salmonella Typhi. Unlike what was observed in the present study, which identified only Salmonella Typhi as the causative agent, no association with other bacterial or viral agents was observed. At this point, analyzes by molecular methods were fundamental for the precise identification of the agent involved, since in the Amazon it is common to have asymptomatic cases of typhoid fever that can evolve to a pulmonary complication easily confused with other conditions existing in the region [13-15,17,18].
Thus, the interaction of Salmonella in cases of gastrointestinal disorders, intestinal perforation, pneumonia, meningitis, vertebral osteomyelitis, septic shock and multiple organ failure seems to be conclusive, but much has been discussed about Salmonella, especially Salmonella Typhi in hepatitis. Thus, studies have linked the occurrence of hepatitis to typhoid fever [18-21]. Although liver cases of typhoid fever can be concomitant with Hepatitis A, there are cases described for cholestatic hepatitis negative for Hepatitis A, B and C [20,21]. Similar to what was observed in the case analyzed by us, which was not observed an association of other agents to the case of cholestatic hepatitis, besides the identification of Salmonella Typhi. Additionally, it was important to note the existence of high genetic similarity between the Salmonella Typhi samples from the three analyzed clinical cases, indicating that although they may be representatives of the same circulating clone, the strains that are generally responsible for typhoid fever, can cause important changes in the host organism without changing the profile of virulence genes frequently involved in classic typhoid fever, and that the analysis of a large number of cases is necessary to unravel the mechanisms that may be involved in complications due to Salmonella infection, in particular Salmonella Typhi.
References
- World health organization, department of vaccines and biologicals (2003) background document: The diagnosis, treatment and prevention of typhoid fever, pp. 19-23.
- Crump JA (2019) Progress in typhoid fever epidemiology. Clin Infect Dis 68(Suppl 1): S4-S9.
- Kumar S, Balakrishna K, Batra HV (2006) Detection of salmonella enterica serovar typhi (s. typhi) by selective amplification of inva, viab, flic-d and prt genes by polymerase chain reaction in multiplex format. Lett Appl Microbiol 42(2): 149-154.
- Das A, Hari SS, Shalini U, Ganeshkumar A, Campus A, et al. (2012) Molecular characterization of salmonella enterica serovar typhi isolated from typhoidal humans. Malaysian Journal of Microbiology 8(3): 148-155.
- Yap KP, Thong KL (2017) Salmonella typhi genomics: Envisaging the future of typhoid eradication. Trop Med Int Health 22(8): 918-925.
- Central for disease control and prevention (CDC) (2020) Typhoid fever and paratyphoid fever-prevention tips for travelers.
- Masinaei M, Eshrati B, Yaseri M (2020) Spatial and spatiotemporal patterns of typhoid fever and investigation of their relationship with potential risk factors in Iran, 2012-2017. Int J Hyg Environ Health 224: 113432.
- Brazil (2020) Ministry of Health (BR). Health Surveillance Secretariat.
- Rocha DCC, Marinho ANR, Reis MSO, Borges IR, Ramos FLP, et al. (2014) Epidemiological profile and molecular characterization of Salmonella Typhi isolated in the State of Pará, Brazil. Pan-Amazonian Journal of Health 5(4): 53-62.
- Townsend SM, Kramer NE, Edwards R, Baker S, Hamlin N, Simmonds M (2001) Salmonella enterica serovar Typhi possesses a unique repertoire of fimbrial gene sequences. Infect Immun 69(5): 2894-2901.
- Levy H, Diallo S, Tennant SM, Tennant SM, Livio S, et al. (2008) PCR method to identify Salmonella enterica serovars Typhi, Paratyphi A, and Paratyphi B among Salmonella Isolates from the blood of patients with clinical enteric fever. J Clin Microbiol 46(5): 1861-1866.
- Baloda SB, Krovacek K, Eriksson L, Linné T, Månsson I (1995) Detection of aerolysin gene in Aeromonas strains isolated from drinking water, fish and foods by the polymerase chain reaction. Comp Immunol Microbiol Infect Dis 18(1): 17-26.
- Casado JL, Navas E, Frutos B, Moreno A, Martin P, et al. (1997) Salmonella lung involvement in patients with HIV infection. Chest 112(5): 1197-1201.
- Wallet F, Pol A, Roussel-Delvallez M, Besson C, Courcol R (1999) Salmonella hadar pneumonia in a heart transplant patient. Clin Microbiol Newslett 21(16): 133-134.
- Ikejiri K, Suzuki K, Ito A, Yasuda K, Shindo A, et al. (2020) Invasive salmonella enteritidis infection complicated by bacterial meningitis and vertebral osteomyelitis shortly after influenza a infection in an immunocompetent young adult. J Infect Chemother 26(2): 269-273.
- Brasil (2010) Ministry of health (BR). Secretariat of health surveillance. department of epidemiological surveillance. integrated manual for surveillance and control of typhoid fever. Ministry of Health, Brazil.
- Jean SS, Tsai MH, Chen WC, Ho SJ, Wu MH, et al. (2009) Fatal community-acquired pneumonia caused by Legionella pneumophila and fluoroquinolone-resistant Salmonella enterica serotype Choleraesuis bacteraemia. Int J Antimicrob Agents 33(2): 189-190.
- Thompson Bastin ML, Neville NR, Parsons RE, Flannery AH, Tennant SJ, et al. (2016) An unusual case of Salmonella enteritidis causing pneumonia, septic shock and multiple organ failure in an immunocompetent patient. ID Cases 6: 85-89.
- El-Newihi HM, Alamy ME, Reynolds TB (1996) Salmonella hepatitis: Analysis of 27 cases and comparison with acute viral hepatitis. Hepatology 24(3): 516-519.
- Shetty AK, Mital SR, Bahrainwala AH, Khubchandani RP, Kumta NB (1999) Typhoid hepatitis in children. J Trop Pediatr 45(5): 287-290.
- Husain EH (2011) Fulminant hepatitis in typhoid fever. J Infect Public Health 4(3): 154-156.
© 2020 Johannes Koch. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






