- Submissions

Full Text
Forensic Science & Addiction Research
Saliva-Blood Adulteration May Impair Age of Stain Determination by RNA Species
Majid Alrowaithi*
Department of Pure and Applied Chemistry, University of Strathclyde, UK
*Corresponding author: Majid Alrowaithi, Department of Pure and Applied Chemistry, University of Strathclyde, Royal College R 6.20, 204 George Street, Glasgow, Scotland, UK
Submission: May 02, 2018;Published: May 16, 2018

ISSN 2578-0042 Volume3 Issue5
Abstract
Analysing the relative quantity of the housekeeping genes β-actin and 18S rRNA has been proposed as genetic method of determining the age of bloodstains collected from crime scenes. However, the measured quantities of these housekeeping genes could be affected in cases of the contamination of bloodstains with another body fluid from the same individual or different individuals. This study investigated the effect of mixing bloodstains with another bodily fluid, namely saliva, on the values of the relative quantity ratio (RQR) between the quantities of β-actin to 18S rRNA and thus on the estimation of the age of the bloodstain. RNA was extracted from fresh and aged blood and saliva separately and in combination with several permutations. RNA was reverse transcripted and quantified for β-actin and 18S rRNA. The data revealed significant changes in the RQR value of β-actin to 18S rRNA in the blood-saliva mixture as compared to separate samples of blood and saliva.
Keywords: Forensic science; Bloodstain; Sliva; Mixture; RNA degradation; Age estimation
Introduction
The determination of the time since the deposition of a biological stain can be crucial to many forensic cases. It may establish a timeframe for when a crime occurred and exclude potential suspects from an investigation. This timestamp becomes especially significant in crimes in which the victim and the suspect are known to one another. Moreover, such estimations can often be used to substantiate other comparable evidence, for instance, witness statements, a suspect’s physical presence, etc.
Although many techniques have been used to determine the age of biological stains, none so far has been developed to a level whereby it can be reliably used in casework. Most research has been conducted on bloodstains, primarily because of their frequent presence [1,2]. These techniques include simple tests that depend on visual observations, as well as those that use sophisticated equipment, such as GC-MS, High-Performance Liquid Chromatography (HPLC) and Electron Paramagnetic Resonance (EPR) spectroscopy [1,3-14]. However, there are many limitations associated with these techniques in terms of precision, accuracy and sample size.
The in vitro instability and lability of RNA drew attention to using it to estimate the time since the deposition of a biological stain. Many studies have been carried out to assess RNA for this purpose. Several published articles have considered the application of RNA to determine blood stain age, with exceptionally promising results [15-17], and have demonstrated the correlation between the age of blood stains and RNA degradation. Anderson et al. [17] have found a statistically significant correlation between the relative quantity ratio (RQR) of β-actin mRNA to 18S rRNA and the time since deposition, or age, of blood stains [16]. The change in the measured RQR of the sample as it aged was attributed to the difference in the degradation rate of the two RNA species, with 18S rRNA being less prone to degradation than β-actin mRNA. It was claimed that this approach offered a number of potential advantages over the previous spectroscopic and chromatographic based techniques. Additionally, this approach could be applied to tissue types other than blood because the RNA species are chosen from genes that are universally expressed. The analysis is not affected by the size of the sample because it examines the RNA ratio [16]. Indeed, Hampton et al. [18] successfully applied this approach to hair samples to determine their age over a period of three months.
However, the markers used in this method are housekeeping RNAs, which are expressed in all tissues at varying levels [19-22]. This may result in changing the RQR value of tested stains in cases in which they have been mixed with other body fluids. This may in turn affect the age estimation of such stains. For example, a mixture of blood with saliva is indistinguishable from only blood because of the consistency of saliva. As the mixture of body fluids, either from one or more people is commonly observed in forensic practice, the aim of this study is to evaluate the effect of blood-saliva mixture on the RQR value of bloodstains and their estimated ages. Four types of mixtures of blood with saliva are considered in this study: fresh blood with fresh saliva, fresh blood with aged saliva, aged blood with fresh saliva and aged blood with aged saliva. Each of these mixtures will be studied as a case. The RQR values were calculated from the ratio of β-actin mRNA over 18S rRNA as suggested by Alrowaithi et al. [23].
Method
Samples
Stains of blood, saliva and a mixture of both were made on clean cotton swabs.
Firstly, pure blood and pure saliva stains were prepared as follows:
Stains made from one type of body fluid, or ‘pure’ stains were prepared from blood and saliva collected from five volunteers, with informed consent, via standard venepuncture for blood and spitting for saliva. From each volunteer, two pure stains for blood (20μL) and two for saliva (50μL) were made on clean cotton swabs to create ten stained swabs of each type of body fluid. Each stain was allowed to dry at room temperature in natural light inside a sterile hood and was then extracted as a dry, fresh, pure stain.
In addition, from blood and saliva collected from four volunteers, eight more pure stains were prepared for each type of body fluid sample from the same volunteers. These additional stains were kept for specific “ages” (i.e., two stains for 10 days, two stains for 30 days and four stains for nine months) before the preparation of mixed stains by the addition of fresh blood or saliva, as described below.
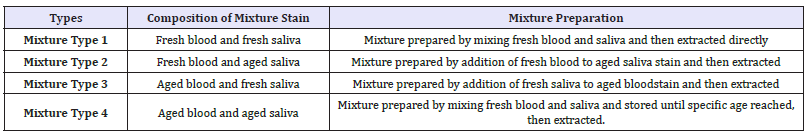
Secondly, four types of mixture stains from the volunteers were prepared from the blood and saliva that had been collected when the stains of pure body fluids were prepared.
The mixture stains consisted of:
1. Fresh blood and fresh saliva were prepared by adding 50μL of fresh saliva to 20μL of fresh aliquoted blood on cotton swabs. These were extracted after drying.
2. Fresh blood and aged saliva were prepared by adding 20μL of fresh blood to previously prepared aged saliva stains.
3. Aged blood and fresh saliva were prepared by adding 50μL of fresh saliva to previously prepared aged bloodstains.
4. Aged stains of blood and saliva were prepared by mixing fresh blood and saliva and stored to reach various ages, as specified above, and left in a dry place until they reached specific ex vivo ages prior to extraction.
All types of mixtures of blood and saliva are shown in Table
1. Each of the following mixture combinations was prepared in duplicate, using blood and saliva from five volunteers for Mixture type 1. Other mixture types were prepared using samples from four volunteers giving 34 mixture stains in all. Figure 1 shows the timeline of collection and preparation of stains prior to extraction.
Figure 1:Timeline of collection and preparation of pure blood and saliva stains and mixture of both of them.
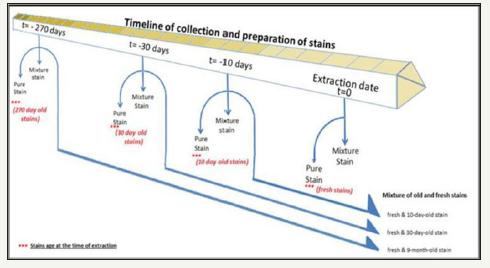
Table 1: Combinations used for mixture study.
RNA extraction
Total RNA was extracted from blood and saliva using an organic extraction method TRI Reagent BD (Molecular Research Center, Cincinnati, Ohio) as stated in manufacturer’s instructions with minor changes.
For each stain, 750μl of TRI Reagent was added to each of the tube containing the stain along with 200μl of water and 3μl of a polyacryl carrier (Molecular Research Center, Cincinnati, Ohio). Tubes were then briefly vortexed and incubated at 50 °C for 10min. One hundred microliters of 1-bromo-3-chloropropane (Molecular Research Center) was added to each tube and then vortexed for 15 sec, followed by incubation at room-temperature for 3min. Tubes were then spun for 15min at 12,000×g at 4 °C. About 500μl of the upper aqueous layer (with the RNA) was transferred to another new tube, and 500μl of cold isopropanal was added. Samples were inverted twice and incubated for 7min at room temperature. The samples were then spun for 8 min at 12,000×g at 4 °C. After discarding the liquid supernatant, 1ml of 75% ethanol was added to wash the RNA pellet. The samples were vortexed and spun for 5min at 12,000×g at 4 °C. After removing the liquid supernatant, the RNA pellets were air dried for 5min at room temperature. Finally, the RNA pellet was re-suspended by addition of 40μl of nuclease-free water followed by incubation at 55 °C for 10min.
Two negative control samples, containing neither blood nor saliva, were treated in the same way. All of the samples were treated with TURBO DNase (Ambion) according manufacturer’s instructions in order to remove contaminating DNA. The RNA quantity was determined spectrophotometrically using a NanoDrop 1000 (NanoDrop Technologies, Wilmington, DE).
Reverse transcription (RT)
Reverse transcription was performed using a Super Script III First-Strand Synthesis System (Invitrogen, Paisley, U.K.). Prior to the actual RT reaction, the random hexamers (Invitrogen), deoxynucleotriphosphate mixture (dNTP) and RNase-free ddH2O were mixed to a final volume of 4μl. Six microliters of RNA sample were then added, and the mixture was incubated for 5min at 65 °C. Samples were chilled on ice immediately and briefly spun before 10× RT buffer (Invitrogen) and Dithiothreitol (DTT) were added. Finally, SuperScript III reverse transcriptase and RNaseOUT were added to each tube, giving a final volume of 20μl that contained 1× RT buffer, 500μM dNTPs, 50ng random hexamers, 5mM MgCl2, 10μM DTT, 200U of SuperScript III and 40U of RNaseOUT. The reaction components were inverted twice and then incubated at room temperature for 10min, followed by incubating at 42 °C for 50min. Finally, the samples were incubated at 70 °C for 15min and then were stored at -20 °C prior to further use.
Quantitative real-time PCR
A real-time PCR master mix was prepared by using primers and probes for the β-actin Reagent Kit Control (cat. No. 401846), Ribosomal RNA control kit (cat. No. 4308329) and a TaqMan Universal PCR Master Mix II (all from Applied Biosystems, Foster City, CA), together with nuclease-free ddH2O. The final concentrations of the reaction were 200nM β-actin probe (FAM dye), 300nM for both the forward and the reverse β-actin primers, 200nM for the 18S rRNA probe (VIC dye), 150nM for both forward and reverse 18S rRNA primers and 1X TaqMan Universal PCR Master Mix. Eighteen microliters of prepared PCR master mix were added to PCR tubes. Two microliters of each cDNA sample were added to the tubes, giving a total volume of 20μl. Duplicate samples were run for each RNA sample, along with a negative control (water) and a positive control (cDNA). ROX dye was included in each sample as a passive reference dye to control for evaporation and pipetting. Samples were amplified in a Mx3005XP qPCR thermocycler (Stratagene) applying the following thermocycle conditions; initial heating at 95 °C for 10min, followed by 40 cycles of 95 °C for 30s, 60 °C for 1min and 72 °C for 1min. Data were analysed using the thermocyclerassociated software. The cycle threshold (Ct value) that was accepted as valid in order to avoid false-positive samples was any value lower than 40.
Statistical analysis
The raw fluorescence Ct values were processed using GenEx statistical software (Version 5.4.0.512; BioEPS GmbH, Munich, Germany) in order to normalize the raw Ct values against interplate calibrators, individual efficiency rates and qPCR technical repeats. Results were tested for a normal distribution and statistically analysed using Minitab® 16 statistical software package (version 16, Minitab Inc., State College, PA, USA).
Results
The statistical evaluation of the RQR values of all samples of all analysed samples, including pure blood, pure saliva and a mixture of blood with saliva, shows that shows that the average RQR value of the mixture differed from that of those composed of only one type of body fluid, i.e., pure samples. Moreover, there is a slight but apparent difference between the RQR values of pure bloodstains and pure saliva stains.
Table 2: Man-Whitney tests carried out on the RQR values of all combination of mixtures between blood and saliva samples, showing the pairwise comparisons of the group means. Asterisk (*) indicates that the difference is significant at the < 0.05 level.
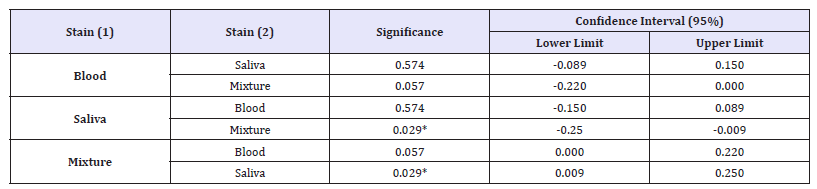
The RQR values were obtained for all samples except those older than 9 months aged stain; the pure blood, the pure saliva stain and the mixtures of them. Interestingly, RQR values were obtained for those 9 months old samples when they mixed with fresh materials.
The results of the statistical analysis performed for all samples strongly pointed to group differences (p-value=0.038). A post hoc comparison, namely a Mann-Whitney test, was performed in order to identify which group differed from the others (Table 2). The result shows that there is a significant difference between the RQR values of mixture samples and those of the pure saliva samples (p-value < 0.05). In addition, no significant difference was found between the RQR values of mixture samples and pure blood samples.
Table 3: Man-Whitney tests carried out on the RQR values of on each type of mixture between bloodstains and saliva stains, showing the pairwise comparisons of the group means.
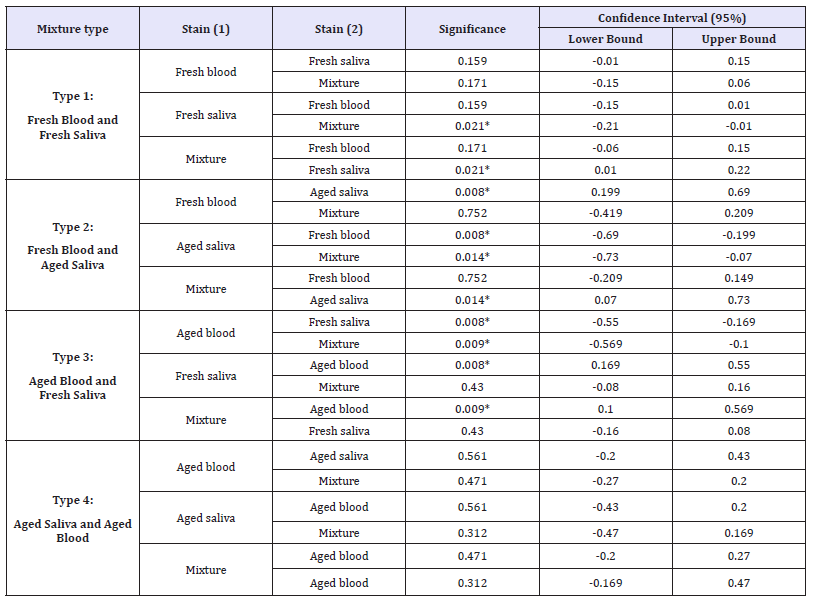
Type 1 is a mixture made of fresh blood and fresh saliva.
Type 2 is a mixture made of fresh blood and aged saliva.
Type 3 is a mixture made of aged blood and fresh saliva.
Type 4 is a mixture made of aged blood and aged saliva. Asterisk (*) indicates that the difference is significant at the < 0.05 level.
The results of the statistical evaluation of each of the four mixture types have shown that the RQR values of some of the mixture types differ from those of pure stains (Table 3). In the first type of mixture (i.e., mixtures of fresh blood and fresh saliva), a significant difference was found between the RQR values of mixture and pure saliva samples (p-value < 0.05). In addition, there was no difference found between the RQR values of either the mixture and pure blood samples or the pure blood and saliva samples (p-value >0.05).
In the second type of mixture (i.e., mixtures of fresh blood and aged saliva), a significant difference was found between aged saliva samples and both mixture and fresh pure blood samples (p-value < 0.01). However, there was no difference found between the RQR values of fresh blood samples and mixture samples (p-value >0.05). Interestingly, it was found that the RQR values of pure saliva stains that are older than 30 days significantly changed when contaminated with fresh blood.
In the third type of mixture (i.e., mixtures of aged blood and fresh saliva), aged blood samples were highly significantly different from both mixture and fresh saliva samples. However, there was no significant difference found between the mixture and the pure saliva samples (p-value >0.05). As in the previous case, the RQR values of pure bloodstains that are older than 30 days significantly changed when contaminated with fresh saliva.
Interestingly, in the fourth type of mixture (i.e., mixtures of aged blood and aged saliva), no significant differences were found between mixture stains and either blood or saliva stains. However, no RQR values were obtained for mixture, blood or saliva stains of more than 9 months old.
Discussion
The method of determining the age of bloodstains developed by Anderson et al. [16] can be considered the most promising molecular method of the determination of the age of a bloodstain collected from a crime scene. Through this method, the age is estimated by measuring the RQR of β-actin mRNA to 18S rRNA. Both of these RNAs are housekeeping RNA genes, which means that they are expressed in all tissues and can be found in all body fluids. This feature was considered to be an advantage because the method can be applied to other tissue types. They will be present in mixtures of different body fluids, such as those found at crime scenes. These mixtures may be from the same individual or different individuals. This study investigated the effect of mixing bloodstains with another bodily fluid, namely saliva, on the RQR values and, in turn, their effect on bloodstain age estimation.
Generally, the statistical analysis of the RQR values of all the stains analysed showed that the RQR values of pure saliva samples differed significantly when contaminated with blood. This may lead to an incorrect estimation of the age of saliva stains in the case of applying this method on saliva stains. However, the effect of such a mixture on the RQR values of pure blood samples was less strong. The change in the RQR values of pure saliva stains when they were contaminated with pure bloodstains may be due to the biological and constitutional differences between them. It has been found that each housekeeping gene is expressed at a different level in each tissue [19,20]. It has also been found that some housekeeping genes show a weaker expression in saliva than in other body fluids. This may explain the lower effect of saliva on the RQR values of bloodstains [24].
However, the results obtained from studying each mixture type separately showed that the effect of one body fluid on the RQR values of the other body fluid depends on the ages of both stains (Figure 2a-2d). In mixture type 1, in which two types of fresh samples were mixed, no significant difference was found between the RQR values of the mixture and those of pure stains, either blood or saliva (Figure 2a). In addition, pure saliva stains show lower RQR values than those of pure blood stains, which may result from the biological and constitutional differences between them. However, this difference was not statistically significant.
Figure 2:RQR values obtained from three stain types (blood, saliva and mixture) in four types of mixtures of blood and saliva demonstrated in a box-and-whiskers plot.
2a: Mixture type 1: a mixture of fresh blood and fresh saliva.
2b: Mixture type 2: a mixture of fresh blood and aged saliva.
2c: Mixture type 3: a mixture of aged blood and fresh saliva.
2d: Mixture type 4: a mixture of aged blood and aged saliva. One outlier was observed for mixture stain type 1.
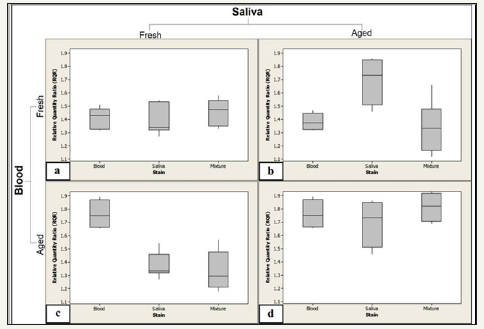
In mixture type 4, in which blood was mixed with saliva and the samples were dried and aged, no significant difference was found between the RQR values of the mixture and those of pure stains, either blood or saliva, or between the RQR values of pure stains themselves (Figure 2d). However, a pure aged saliva stain exhibits a larger diversity in RQR values as compared to those of pure bloodstains. This large range may be due to the constitutional components of saliva. It is known that saliva contains various endoand exo-ribonucleases from many sources, e.g., viral and bacterial ribonucleases, as well as extraneous substances, such as food debris [25,26]. These substances have different effects on RNA integrity in saliva, which may result in the wide range of RQR values observed.
The greatest effect of mixture on RQR value appeared in mixture types 2 and 3, in which old samples were mixed with a fresh sample of a different type (Figure 2b & 2c). It was found that the RQR values of pure stains that were older than a month differed significantly when mixed with fresh samples. Moreover, this effect appeared clearly in samples older than 9 months: the results changed from an inability to obtain an RQR value in pure samples to obtaining an RQR value when a fresh sample of either blood or saliva was added to an old, i.e., more than 9 months old, stain.
Conclusion
In conclusion, although using housekeeping RNA molecules has been considered an advantage of the method developed by Anderson et al. [16], it could be a limitation in a case in which two types of body fluids were mixed. The results of this study show the effect of the contamination of bloodstains with saliva on RQR values and in turn on their estimated ages. A marked effect of fresh saliva on the RQR values of old bloodstains and a marked effect of fresh blood on the RQR values of old saliva stains were noticed, which demonstrates that the effect of a mixture on the RQR value resulted from the difference in the ages of the stains. The amount of body fluid in the mixed samples may also have an effect on the RQR value of a mixed stain, and this should be studied further. Other body fluids, such as semen and sweat, may have the same effect on the RQR values of bloodstains, and these should be investigated as well.
This work used the method developed by Anderson et al. [16], which is considered to be the most promising molecular method of bloodstain age determination. However, this work revealed difficulties in the use of housekeeping gene RNA for the purpose of establishing the age of bloodstains that have been mixed with other bodily fluids, whether from the same person or another person. This is a common situation in forensic practice [27]. Therefore, the use of housekeeping gene RNA to determine the age of bloodstains or other body fluids should be accompanied by a body fluid identification in order to identify mixtures. As the source of the stains in forensic casework will, by definition, be unknown, the underlying assumptions concerning the composition of the stains must be made clear. The use of tissue-specific genes instead of housekeeping genes to determine the age of bloodstains could be a solution to the effect of mixtures with other body fluids.
Acknowledgement
The present study was supported by a grant from the General Directorate of Public Security of the Kingdom of Saudi Arabia. The authors are grateful to Dr Eve Lutz for welcoming us to SIPBS and her invaluable support.
References
- Bremmer RH, de Bruin KG, Gemert MJ, Leeuwen TG, Aalders MCG (2012) Forensic quest for age determination of bloodstains. Forensic Sci Int 216(1-3): 1-11.
- James SH, Kish PE, Sutton TP (2005) Principles of bloodstain pattern analysis: theory and practice, CRC Press, Boca Raton, USA
- Fiori A (1962) Detection and identification of bloodstains, in the methods of forensic science. Interscience, New York, USA, pp. 243-290.
- Inoue H, Takabe F, Iwasa M, Maeno Y (1991) Identification of fetal hemoglobin and simultaneous estimation of bloodstain age by highperformance liquid chromatography. International Journal of Legal Medicine 104(3): 127-131.
- Inoue H, Takabe F, Iwasa M, Maeno Y, Seko Y (1992) A new marker for estimation of bloodstain age by high performance liquid chromatography. Forensic Sci Int 57(1): 17-27.
- Lins G, Blazek V (1982) The use of remission analysis for direct colorimetric determination of age of blood stains. J Legal Med 88(1-2): 13-22.
- Shinomiya T, Muller M, Muller PH, Lesage R (1978) Immunoelectrophoresis used for identification of blood stains in forensic medicine. Forensic Sci Int 12(2): 157-163.
- Kind SS, Watson M (1973) The estimation of blood stain age from the spectrophotometric properties of ammoniacal blood stain extracts. Forensic Sci Int 2(3): 325-332.
- Bremmer RH, Nadort A, Leeuwen TG, Gemert MJ, Aalders MC (2010) Age estimation of blood stains by hemoglobin derivative determination using reflectance spectroscopy. Forensic Sci Int 206(1-3): 166-171.
- Andrasko J (1997) The estimation of age of bloodstains by HPLC analysis. Journal of Forensic Sciences 42(4): 601-607.
- Matsuoka T, Taguchi T, Okuda J (1995) Estimation of bloodstain age by rapid determinations of oxyhemoglobin by use of oxygen electrode and total haemoglobin. Biological & Pharmaceutical Bulletin 18(8): 1031.
- Fujita Y, Tsuchiya K, Abe S, Takiguchi Y, Kubo S, et al. (2005) Estimation of the age of human bloodstains by electron paramagnetic resonance spectroscopy: long-term controlled experiment on the effects of environmental factors. Forensic Sci Int 152(1): 39-43.
- Strasser S, Zink A, Kada G, Hinterdorfer P, Peschel O, et al. (2007) Age determination of blood spots in forensic medicine by force spectroscopy. Forensic Sci Int 170(1): 8-14.
- Arany S (2011) Age estimation of bloodstains: A preliminary report based on aspartic acid racemization rate. Forensic Sci Int 212(1-3): e36-e39.
- Bauer M, Polzin S, Patzelt D (2003) Quantification of RNA degradation by semi-quantitative duplex and competitive RT-PCR: a possible indicator of the age of bloodstains? Forensic Sci Int 138(1-3): 94-103.
- Anderson S, Howard B, Hobbs GR, Bishop CP (2005) A method for determining the age of a bloodstain. Forensic Sci Int 148(1): 37-45.
- Anderson SE, Hobbs GR, Bishop CP (2011) Multivariate analysis for estimating the age of a bloodstain. Journal of Forensic Sciences 56(1): 186-193.
- Hampson C, Louhelainen J, McColl S (2011) An RNA expression method for aging forensic hair samples. Journal of Forensic Sciences 56(2): 359- 365.
- Barber RD, Harmer DW, Coleman RA, Clark BJ (2005) GAPDH as a housekeeping gene: analysis of GAPDH mRNA expression in a panel of 72 human tissues. Physiological Genomics 21(3): 389-395.
- Hsiao LL, Dangond F, Yoshida T, Hong R, Jensen RV, et al. (2001) A compendium of gene expression in normal human tissues. Physiological Genomics 7(2): 97-104.
- Moreno LI (2012) Determination of an effective housekeeping gene for the quantification of mRNA for forensic applications. Journal of Forensic Sciences 57(4): 1051-1058.
- Li Y (2004) RNA profiling of cell-free saliva using microarray technology. Journal of Dental Research 83(3): 199-203.
- Alrowaithi MA, McCallum NA, Watson ND (2014) A method for determining the age of a bloodstain. Forensic Science International 234: e30-e31.
- Haas C, Klesser B, Maake C, Bär W, Kratzer A (2009) mRNA profiling for body fluid identification by reverse transcription endpoint PCR and realtime PCR. Forensic Sci Int: Genetics 3(2): 80-88.
- Park NJ, Li Y, Yu T, Brinkman B, Wong DT (2006) Characterization of RNA in saliva. Clinical Chemistry 52(6): 988.
- Eichel HJ, Conger N, Chernick WS (1964) Acid and alkaline ribonucleases of human parotid, submaxillary, and whole saliva. Archives of Biochemistry and Biophysics 107: 197-208.
- Butler J (2005) Forensic DNA typing: biology and technology behind STR markers, (2nd edn), Academic Press, London, UK.
© 2018 Majid Alrowaithi. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






