- Submissions

Full Text
Examines in Marine Biology & Oceanography
Potential Use of Marine Compounds to Modulate the Ubiquitin-Proteasome System
Stefano Magnati1, Leonardo Mortati2 and Enrico Bracco2,3*
1Anti Doping Center, A. Bertinaria, San Luigi Hospital, Orbassano, Italy
2Istituto Nazionale di Ricerca Metrologica, Torino, Italy
3Department of Oncology, University of Turin, Italy
*Corresponding author: Enrico Bracco, Department of Oncology, Torino, Italy
Submission: June 19, 2023;Published: June 28, 2023

ISSN 2578-031X Volume6 Issue1
Abstract
The Ubiquitin-Proteasome System (UPS) is a major intracellular protein degradation system, highly conserved throughout life kingdoms, where the proteasome is the core. Due to its enhanced or inhibited activity, dysregulation of the UPS has undoubtedly been implicated in the pathogenesis of various diseases, including cancer and neurodevelopmental and neurodegenerative disorders. Consequently, given its crucial role in maintaining cellular homeostasis and protein quality control, targeting the different UPS components has emerged as a promising therapeutic strategy. Here, we shortly review the capability of marine-derived compounds as novel modulators of the proteasome under their peculiar features, making them attractive for future therapeutic applications. Marine compounds present in disparate organisms inhabiting oceans exhibit different biological activities and often possess specific proteasome-modulating properties. Currently, dozens of newly isolated marine-derived compounds have shown the ability to modulate proteasome activity at very low concentrations (e.g., nano- to the micro-molar range), pointing out their superior potency and efficacy and envisaging a potential future use in the clinic. Nowadays, the marine environment has represented a fruitful “spring” for identifying different unique metabolites, and to several extents, arguably, it still holds overwhelming scientific wonders to modulate the UPS.
Keywords:Marine compounds; Proteasome system; Neurodevelopmental; Neurodegenerative disorders; Cellular homeostasis; Therapeutic strategy; Peculiar features
Introduction
Proteins are responsible for nearly every cellular function. On average, a human cell expresses 10,000-13,000 different protein species [1]. Hence, a huge network of factors is committed to preserving protein homeostasis (proteostasis) and preventing the assembly and accumulation of potentially toxic protein aggregates. Therefore, protein synthesis and degradation must be finely tuned. Malfunctioning proteins, either because they are damaged or abnormally folded, as well as regulatory proteins, such as cell-cycle, cell-signaling, and apoptosis regulators, are disposed of by various means, including the Ubiquitin-Proteasome System (UPS), whose core is the proteasome machinery [2]. The latter machinery is structurally organized as a multi-subunit and multi-catalytic protease localized at the cytosolic as well as at nuclear levels. The proteasome consists of a barrel-shaped proteolytic core with an axial cavity (20S proteasome), which contains four stacked rings. The stacked heptameric rings include two outermost non-catalytic α-rings and two innermost catalytic β-rings arranged in a αββα structure [3] (Figure 1A). In addition to the central processing unit (20S) there is a regulatory particle (19S), exhibiting ATPase activity, which is actively implied in the protein unfolding and in the removal of the ubiquitin moiety (chain/s) prior the entering of the protein into the central cavity of the 20S and its following degradation into small peptides. When a 19S unit caps, the 20S core gives rise to a 26S proteasome conformation, while when both sides of the core barrel are occupied by the 19S, a 30S proteasome is assembled. Usually, proteins destined for proteasomal degradation are tagged by polyubiquitin (Ub) chain/s (especially with Lys-48 linkages).
The protein ubiquitination is a stepwise, evolutionary, deeply conserved process that implies the sequential involvement of three different enzymes [4]. E1 activates Ub in an ATP-dependent fashion. Upon activation, Ub is then transferred to an E2-conjugating enzyme, which ultimately, with the assistance of the E3-Ub ligases, transfers the Ub moiety to the substrate protein. It is not surprising that the dysregulation of the UPS, either because the proteasome activity is impaired or because augmented, is associated with several human diseases, including solid and hematological malignancies. Intriguingly, proliferating tumor cells are more sensitive to proteasome inhibition when compared to non-proliferating cells [5], suggesting that proteasome activity is required to sustain the neoplastic phenotype by promoting proteotoxicinduced detoxification. Therefore, inhibiting proteasome leads to proteotoxic stress that sensitizes cancerous cells toward apoptosis. Conversely, among aging hallmarks is the age-related deterioration of the proteostasis network. Indeed, the proteasome function declines significantly upon aging, contributing to the manifestation of age-related pathologies, such as neurodegenerative disorders [6]. Interestingly, UPS dysregulation has also been associated with neurodevelopmental disorders [7]. Hence, the possibility to modulate the proteasome activity has been seen as an attractive tool to be exploited, and many attempts have been undertaken toward proteasome activation as a strategy to promote health span and longevity, or conversely toward its inhibition for neurodegenerative disorders and cancers, respectively (Figure 1A). The marine environment hosts a myriad of organisms that might potentially act as an excellent reservoir of primary and secondary metabolites, which exhibit a wide spectrum of biological activities.
Figure 1:A) The upper part represents neurodegenerative disorders characterized by impaired proteasome activity,
emphasizing the potential therapeutic strategy of proteasome activation. The lower part showcases a general cancerous
cell with elevated proteasome activity, suggesting that the proteasome is a valuable target for inhibition in cancer
therapy.
B) Barplots depict the increasing scientific interest and research on proteasome activators and inhibitors over the
years. The data was obtained from PubMed using the keywords ‘Proteasome activator’ and ‘Proteasome inhibitor’.
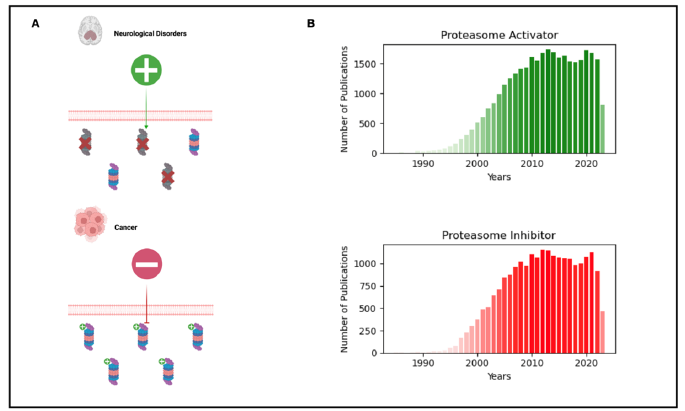
Remarkably, in the last decades, we have witnessed that several marine compounds display potential (direct or indirect) actions on UPS components, including the proteasome, thus serving as UPS modulators and exerting beneficial effects on conditions such as oxidative stress, aging, and age-related disorders. To date, the main sources of marine bioactive products displaying the properties of modulating the proteasome activity are represented by sponges and their associated microbial communities. However, marine invertebrates such as soft corals and tunicates might represent a fruitful source of active biomolecules suitable for modulating proteasome activity.
Marine-Derived Compounds with Proteasome- Inhibiting and -Enhancing Properties
In the last decades, as witnessed by the large amount of data accumulated, we have witnessed an increasing interest in the scientific community in attempting to identify compounds displaying proteasome-modulating effects (Figure 1B). Cancerous cells significantly rely on proteasomal activity, which is tightly associated with their robust proliferative and metabolic rates sustained by enhanced protein synthesis. Hence, a tight protein turnover involving the proteasome is required. The precursor of all marine-derived natural products acting as proteasome modulators is Salinosporamide A (Marizomib) which exhibits peculiar inhibitory features [8]. Compared to the previous proteasome inhibitor Bortezomib, Salinosporamide A displays fewer side effects, and it is currently under investigation for hematological (i.e., Multiple Myeloma) and Central Nervous System tumors [9]. Indeed, diversly from many analogous compounds (i.e., Bortezomib, Carfilzomib, and Ixazomib) it is an irreversible proteasome inhibitor that is blood-brain barrier permeant. After that, several bioactive molecules displaying inhibitory effects on the proteasome, in the range of nano to micro-molar concentrations, have been isolated and characterized in cellular and animal models. Basically, to date such inhibitors fall into four main chemical categories: a) alkaloids (e.g., mazamines, oroidin-derivatives, variabines) [10-12]; b) terpenes (e.g., strongylophorine, heteronemin, petrosaspongiolide) [13-15]; c) quinones (e.g., halenaquinones derivatives) [16], and d) peptides (e.g., fellutamide, carmaphycin, and scytonemide) [17-19]. The proteasome proteolytic activity is achieved with different cleavage preferences, caspase-like, trypsin-like, and chymotrypsin-like performed by the different β subunits, β1, β2, and β5 respectively [3]. Interestingly, quite often marine-derived compounds with proteasome-inhibiting activities display different site-specific inhibition enabling them to selectively inhibit the different cleavage preferences.
Conversely from cancerous cells, neurodegenerative disorders are characterized by the accumulation of toxic intracellular protein aggregates (e.g., Parkinson, Alzheimer’s disease) mostly due to a reduced proteasome activity. Therefore, enhancing the activity of the latter by small-molecule compounds has been assumed to be a valuable approach in treating neurodegenerative disorders [6,20]. Differently from most bioactive marine-derived proteasome inhibitor compounds that directly interact and thus inhibit the proteasome, those that enhance proteasome activity exert their action in an indirect fashion. Indeed, they determine an increase in the cellular proteasomal level through transcriptional induction of proteasomal subunit encoding genes. Among these compounds, sulforaphane [21], phenolic compounds such as phloroglucinol [22-24], astaxanthin, fucoxhantin [25], and benzo-1,3-oxazine xyloketal derivative [26,27] have displayed, to different extents, a neuroprotective and antioxidant functions in cellular and animal models.
Conclusion and Future Perspectives
Though proteasome inhibition is a promising therapeutical approach, it is currently facing a few hurdles. Indeed, such compounds display serious side effects that restrict their applicability and beneficial effects. In addition, in patients affected by hematological malignancies (e.g., multiple myeloma), after an initial response, such molecules lose their efficacy because of the development of drug resistance mechanisms. Additionally, solid tumors display primary resistance to the currently available proteasome inhibitors. In this regard, marine compounds might represent a valuable alternative to be exploited. Remarkably, carphamycins exhibit good efficacy against solid tumors [18]. As future challenges, the pharmacological and preclinical assessment of proteasome inhibitors marine-derived compounds, either used as a single agent or in combination with the classic ones (e.g., Bortezomib) or with other drugs other than proteasome inhibitors, should be assessed in view to sensitize the pharmaco-resistance. Furthermore, since proteasomes are common to archebacteria, mycobacteria (e.g., Mycobacterium tuberculosis), and eukaryotes it would be of great interest to appraise the activity of marine-derived proteasome inhibitor molecules also in these cells due to the active involvement of the proteasome machinery in the pathogenesis of the associated diseases (e.g., tuberculosis, malaria, leishmaniasis, Chagas disease, etc..). As far as it concerns the marine-derived proteasome activators, we are lagging a little bit behind.
In the near future, the most challenging requirement is to gain deeper insights into their in vivo effects on mammalian model organisms (e.g., murine) and provide preclinical data to support further clinical phase I/II clinical trials potentially. Albeit the marine milieu represents a fruitful “spring” of metabolites where to draw, there are some serious practical drawbacks, including the yield of the extracted molecules and the ecosystem menacing. To this purpose, the development of chemical synthesis and the genetically modified system might be helpful to remedy such issues.
References
- Bekker-Jensen DB, Kelstrup CD, Batth TS, Larsen SC, Haldrup C, et al. (2017) An optimized shotgun strategy for the rapid generation of comprehensive human proteomes. Cell Systems 4(6): 587-599.
- Schwartz AL, Ciechanover A (2009) Targeting proteins for destruction by the ubiquitin system: Implications for human pathobiology. Annu Rev Pharmacol Toxicol 49: 73-96.
- Bard JAM, Goodall EA, Greene ER, Jonsson E, Dong KC, et al. (2018) Structure and function of the 26S proteasome. Annu Rev Biochem 87: 697-724.
- Pergolizzi B, Bozzaro S, Bracco E (2019) Dictyostelium as model for studying ubiquitination and deubiquitination. Int J Dev Biol 63(8-9-10): 529-539.
- An B, Goldfarb RH, Siman R, Dou QP (1998) Novel dipeptidyl proteasome inhibitors overcome bcl-2 protective function and selectively accumulate the cyclin-dependent kinase inhibitor P27 and induce apoptosis in transformed, but not normal, human fibroblasts. Cell Death Differ 5(12): 1062-1075.
- Dantuma NP, Bott LC (2014) The ubiquitin-proteasome system in neurodegenerative diseases: Precipitating factor, yet part of the solution. Front Mol Neurosci 7: 70.
- Ebstein F, Küry S, Papendorf JJ, Krüger E (2021) Neurodevelopmental Disorders (NDD) caused by genomic alterations of the Ubiquitin-Proteasome System (UPS): The possible contribution of immune dysregulation to disease pathogenesis. Front Mol Neurosci 14: 733012.
- Potts BC, Albitar MX, Anderson KC, Baritaki S, Berkers C, et al. (2011) Marizomib, a proteasome inhibitor for all seasons: Preclinical profile and a framework for clinical trials. Curr Cancer Drug Targets 11(3): 254-284.
- Boccellato C, Kolbe E, Peters N, Juric V, Fullstone G, et al. (2021) Marizomib sensitizes primary glioma cells to apoptosis induced by a latest-generation TRAIL receptor agonist. Cell Death Dis 12(7): 647.
- El-Desoky AH, Kato H, Eguchi K, Kawabata T, Fujiwara Y, et al. (2014) Acantholactam and pre- neo -kauluamine, manzamine-related alkaloids from the indonesian marine sponge acanthostrongylophora ingens. J Nat Prod 77(6): 1536-1540.
- Sakai E, Kato H, Rotinsulu H, Losung F, Mangindaan REP, et al. (2014) Variabines A and B: New β-carboline alkaloids from the marine sponge luffariella variabilis. J Nat Med 68(1): 215-219.
- Lansdell TA, Hewlett NM, Skoumbourdis AP, Fodor MD, Seiple IB, et al. (2012) Palau’amine and related oroidin alkaloids dibromophakellin and dibromophakellstatin inhibit the human 20S proteasome. J Nat Prod 75(5): 980-985.
- Noda A, Sakai E, Kato H, Losung F, Mangindaan REP, et al. (2015) Strongylophorines, meroditerpenoids from the marine sponge petrosia corticata, function as proteasome inhibitors. Bioorganic & Medicinal Chemistry Letters 25(13): 2650-2653.
- Schumacher M, Cerella C, Eifes S, Chateauvieux S, Morceau F, et al. (2010) Heteronemin, a spongean sesterterpene, inhibits TNF α-induced NF-ΚB activation through proteasome inhibition and induces apoptotic cell death. Biochemical Pharmacology 79(4): 610-622.
- Margarucci L, Monti MC, Tosco A, Riccio R, Casapullo A (2010) Chemical proteomics discloses petrosapongiolide m, an antiinflammatory marine sesterterpene, as a proteasome inhibitor. Angewandte Chemie International Edition 49(23): 3960-3963.
- Tsukamoto S, Yamakuma M, Kato H, Matsuo K, El-Desoky A, et al. (2014) 1-hydroxyethylhalenaquinone: A new proteasome inhibitor from the marine sponge xestospongia Sp. Heterocycles 89(11): 2605.
- Pirrung MC, Zhang F, Ambadi S, Gangadhara Rao Y (2016) Total synthesis of fellutamides, lipopeptide proteasome inhibitors. More sustainable peptide bond formation. Org Biomol Chem 14(35): 8367-8375.
- Pereira AR, Kale AJ, Fenley AT, Byrum T, Debonsi HM, et al. (2012) The carmaphycins: New proteasome inhibitors exhibiting an α,β-epoxyketone warhead from a marine cyanobacterium. Chem Bio Chem 13(6): 810-817.
- Krunic A, Vallat A, Mo S, Lantvit DD, Swanson SM, et al. (2010) Scytonemides A and B, cyclic peptides with 20s proteasome inhibitory activity from the cultured cyanobacterium scytonema hofmanii. J Nat Prod 73(11): 1927-1932.
- Lee BH, Lee MJ, Park S, Oh DC, Elsasser S, et al. (2010) Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature 467(7312): 179-184.
- Liu Y, Hettinger CL, Zhang D, Rezvani K, Wang X, et al. (2014) Sulforaphane enhances proteasomal and autophagic activities in mice and is a potential therapeutic reagent for huntington’s disease. J Neurochem129(3): 539-547.
- Ryu J, Zhang R, Hong BH, Yang EJ, Kang KA, et al. (2013) Phloroglucinol attenuates motor functional deficits in an animal model of parkinson’s disease by enhancing Nrf2 activity. Plos One 8(8): e71178.
- Yang EJ, Mahmood U, Kim H, Choi M, Choi Y, et al. (2018) Phloroglucinol ameliorates cognitive impairments by reducing the amyloid β Peptide burden and pro-inflammatory cytokines in the hippocampus of 5XFAD mice. Free Radical Biology and Medicine 126: 221-234.
- Yang EJ, Ahn S, Ryu J, Choi MS, Choi S, et al. (2015) Phloroglucinol attenuates the cognitive deficits of the 5XFAD mouse model of Alzheimer’s Disease. PLoS ONE 10(8): e0135686.
- Zhang L, Wang H, Fan Y, Gao Y, Li X, et al. (2017) Fucoxanthin provides neuroprotection in models of traumatic brain injury via the Nrf2-ARE and Nrf2-autophagy pathways. Sci Rep 7: 46763.
- Zeng Y, Guo W, Xu G, Feng L, Long S, et al. (2016) Xyloketal-derived small molecules show protective effect by decreasing mutant huntingtin protein aggregates in Caenorhabditis Elegans Model of Huntington’s disease. DDDT 10: 1443-1451.
- Zhou JB, Zheng YL, Zeng YX, Wang JW, Pei Z, et al. (2018) Marine derived xyloketal derivatives exhibit anti-stress and anti-ageing effects through HSF pathway in caenorhabditis elegans. European Journal of Medicinal Chemistry 148: 63-72.
© 2023 Enrico Bracco. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






