- Submissions

Full Text
Examines in Marine Biology & Oceanography
Seagrass Omics: How to Short the Workflow for Protein Expression Analyses
Daniela Oliva, Amalia Piro*, Matteo Nisticò D, Faustino Scarcelli and Silvia Mazzuca
Laboratory of Plant Biology and Plant Proteomics (Lab.Bio.Pro.Ve.), Department of Chemistry and Chemical Technologies, Università della Calabria, 87036 Rende, Italy
*Corresponding author: Amalia Piro, Laboratory of Plant Biology and Plant Proteomics (Lab.Bio.Pro.Ve.), Department of Chemistry and Chemical Technologies, Università della Calabria, 87036 Rende, Italy
Submission: February 02, 2023;Published: February 28, 2023

ISSN 2578-031X Volume5 Issue4
Abstract
When planning a differential protein expression trial in seagrass populations, time consuming and expensive workflow is the main issue to be addressed. Here we reported a faster procedure from the electrophoretic separation to the mass spectrometric analysis of Posidonia oceanica leaf proteins, selected among three different acrylamide/diacrylamide concentrations, three different running times of denaturing electrophoresis followed by in-gel digestion reactions, three total mass spectrometry run times. We demonstrated that the procedure using 12 or 10% polyacrylamide gel, the SDS-PAGE duration of 15 min and the total LC-MS/MS run of 120 min, did not affect the proteolytic efficiency of trypsin, improved the quality of the generated peptides, increased the number of identified proteins belonging to many functional categories per sample, compared to those of a standard workflow and of the other tested conditions.
Keywords:Seagrasses; Proteomics; In-gel digestion; SDS-PAGE; Mass spectrometry
Introduction
Since the last decade, proteomic technologies have been successfully applied to marine plants to obtain insights into their adaptive capacity and gene expression plasticity against environmental threats [1-7]. In seagrasses several species-specific multistep workflows for free-label differential protein expression have been optimized to obtain high quality protein samples [8-10]; these protocols were routinely coupled with the tryptic in-gel digestion as this method offers the advantage to remove any further contaminants (e.g., detergents, salts) during electrophoresis. The generated peptides can be, then, readily analysed by LC-MS/MS to obtain the peptide mass fingerprinting. The main drawback of in-gel digestion is that a single SDS-PAGE gel lane, corresponding to a biological or technical replicate, is excised, divided in several slices and splitted in several independent digestion reactions [11] and thus in multiple mass spectrometry analyses. This is a time lengthly, costly procedure and represents the main technical issue to be faced in the differential protein expression trials. In order to improve and encourage the use of the proteomic approach to investigate at molecular level the seagrass biology and adaptation strategies, here we proposed a workflow that used a short-term electrophoresis in which protein sample was concentrated in lane of one centimeter long instead of tens; in our experience, the short-term SDS-PAGE required a single digestion reaction per sample and significantly reduced time and cost of the entire trial. Here we investigated the effects of the shortened duration of the eletrophoresis on the proteolytic efficiency, the quality of generated peptides and the number of identified proteins. Results were also discussed in terms of functional classification of the identified proteins using the Pather.db classification tools [12].
Material and Methods
Cuttings of P. oceanica were collected by scuba divers in a large meadows along the coast of Calabria (Souther Italy) (39,697216; 15,803951, February 2022). Leaves of each shoot have been cleaned of epiphytes, washed quickly in water, frozen in liquid N2 and stored at -80 °C. The leaf tissues were used for protein extraction as described in Piro et al. [4]. Briefly, 1gr of frozen leaves were ground in a mortar untill a fine tissue powder was obtained. The powder was divided into 2ml vials; a volume of 10% TCA in acetone was added and centrifuged at 13000rpm for 5 min at 4 °C. The pellet containing the precipitated proteins was whased in 80% acetone and dried at room temperature. For orotein extraction of proteins, 0.1g of pellet was dissolved in 0.8ml of phenol (buffered withTris- HCL,pH8.0,Sigma,St. Louis, MO, USA) and 0.8ml of SDS buffer (30% sucrose, 2% SDS,0.1M Tris-HCl, pH8.0, 5% 2-mercaptoetanol) in 2ml vials. After centrifugation the phenol phase was added with five volumes of 0.1M ammoniumacetate in cold methanol, and the mixture was stored at -20 °C for 30 min to precipitate proteins. Proteins were collected by centrifugation at 13000rpm for 5 min and washed in 0,1M ammoniumacetate/methanol and 80% acetone.The final pellet containing purified proteins were dried at room temperature and dissolved at final concentration with Laemli 1D electrophoresis buffer [13]. Proteins were then quantified by the bradford assay. Protein yield was measured as mg of protein per g fresh tissue weight in three independent biological replicates for each test.
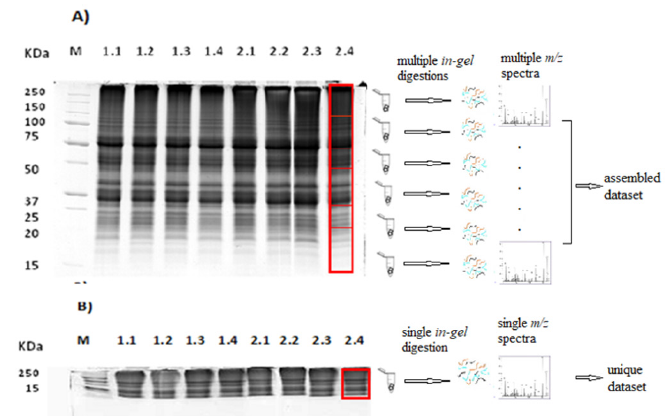
Purified protein samples were laoded on three gels of 6%, 10% and 12% polyacrylamide; short-term SDS-PAGEs were runned for 15 min, while control electrophoresis was runned for 1h and 15 min on 12% acrylamide gel at same run conditions [13]. In this way, three biological replicates for each condition have been runned; after short-term electrophoresis each replicate resulted in one large band and it has been processed as a single in-gel digestion reaction. As control, same replicates were separated in a classical eletrophoresis and each lane was splitted in six in-gel digestion vials (Figure 1). Mass spectra were obtained by an EASYLC 1000 coupled to a Q-Exative Classic Benchtop Quadrupole- Orbitrap hybrid Mass Spectrometer (Thermo Fisher Scientific, Germany) at three different total run times of 45 min, 60 min and 120 min. Each control vial has been analysed at 60 min run time, resulting in 360 min total time duration per replicate. Protein inference and validation were performed with Scaffold software 4.8, unsig a setting with statistical parameters of highest stringency (Proteome Software, Inc., Portland, USA). Through the X!Tandem software peptides in samples were aligned and identified against Zostera marina genome sequences deposited in the NCBI databases (dowloaded on November 2022).
Figure 1:12% polyacrilammide SDS-PAGEs of leaf proteins separated by A) A classic run in which lanes had a length of 10cm. Each lane was cut in six slices and processed via a typical in-gel digestion protocol followed by a multiple mass spectrometry workflow each of 60 min generating six spectra datasets that have to be assembled ; B) The shorten run in which lanes had a length of approx 1,5cm. Each lane was then cut as a single sample for processing as a single in-gel digestion, as a single mass spectrometry workflow of 120 min generating a unique spectra dataset.
Result and Discussion
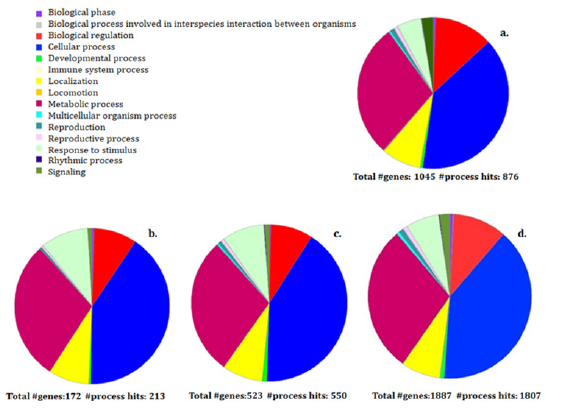
Short-term SDS-PAGE Procedure using 12 or 10% acrylammide gels, 15 min run SDS-PAGE, 120 min LC-MS/MS total run time, setting at least five exclusive peptides per protein as threshold for identification significance, gave higher number of identified proteins (1977±130) respect to the 1h 15 min run replicates (1143±60), the 60 min run replicates (552±55) and 45 min run replicates (193±25). Functional classification of proteins made by Panther.db tool [12] from the 120 min method resulted in 19 Protein classes belonging to 15 Biological processes in respect to 17 and 13 categories in control method respectively; the catergories overlapped for 70% and 86% respectively between two methods (Figure 2). Based on our results, the use of short-term gel electrophoresis improved sample processing; in fact, it did not affect tryptic digestion efficiency but generated peptides that resulted in a higher number of proteins identified than those of a classical workflow. This result suggests that within the band in short-term gel there are more proteins with very low molecular weight that, generally, could be lost during a longer electrophoretic run. The utility of short-term electrophoresis, in addition to immobilizing proteins to take advantage of gel digestion, minimizes gel length and allows to process a single digestion reaction per sample and avoid multiple runs in the mass spectrometry workflow.
Figure 2:Functional classification by PANTHER analysis of the biological processes of the identified proteins from regular SDS-PAGE duration (a) and from short-term 12% polyacrilammide gel eletrophoresis after 45 min (b) 60 min (c) and 120 min (d) of total mass spectrometry times.
Conclusion
In our work, short-term electrophoresis coupled with a single in-gel tryptic digestion reaction reduced the time and cost required for protein separation and mass spectrometry analysis by seventy percent. The seagrass research community is strongly committed to linking ecophysiology with molecular approaches to elucidate plant responses against different environmental conditions [14- 16]. Seagrass proteomics is currently able to assign significant interpretations to an increasing number of differentially expressed genes; in this context, our fine-tuned technique will allow to analyze a larger statistical sample, without having to undergo very long and expensive workflows. This result could make proteomic specialty much more feasible in differential protein expression studies of seagrass populations.e “less to lose” and hence are capable of taking higher risks [40,41].
Acknowledgment
The authors thanks the PON, Grant number a3_00341A S.I.L.A.- Integrated System of Environmental Laboratories-University of Calabria for mass spectrometry equipments. This research was financed with a PhD fellowship from Università della Calabria, High Education Resources funds 2021-2024.
References
- Piro A, Bernardo L, Serra IA, Barrote I, Olivé I, et al. (2020) Leaf proteome modulation and cytological features of seagrass Cymodocea nodosain response to long-term high CO2 exposure in volcanic vents. Sci Rep 10(1):
- Procaccini G, Ruocco M, Marín-Guirao L, Dattolo E, Brunet C, et al. (2017) Depth-specific fluctuations of gene expression and protein abundance modulate the photophysiology in the seagrass Posidonia oceanica. Sci Rep 7:
- Kumar M, Padula MP, Davey P, Pernice M, Jiang Z, et al. (2017) Proteome analysis reveals extensive light stress-response reprogramming in the seagrass Zostera muelleri (Alismatales, Zosteraceae) Frontiers in Plant Science 7: 2023.
- Piro A, Marín-Guirao L, Serra IA, Spadafora A, Sandoval-Gil JM, et al. (2015) The modulation of leaf metabolism plays a role in salt tolerance of Cymodocea nodosa exposed to hypersaline stress in mesocosms. Frontiers in Plant Science 6: 464.
- Dattolo E, Gu J, Bayer P, Mazzuca S, Serra IA, et al. (2013) Acclimation to different depths by the marine angiosperm Posidonia oceanica: Transcriptomic and proteomic profiles. Frontiers in Plant Science 4: 195.
- Mazzuca S, Filadoro D, Vannini C, Marsoni M, Cozza R, et al. (2009) Seagrass light acclimation: 2-DE protein analysis in Posidonia leaves grown in chronic low light conditions. J Exp Mar Biol Ecol 374(2): 113-122.
- Spadafora A, Filadoro D, Mazzuca S, Bracale M, Marsoni M, et al. (2008) 2-DE polypeptides mapping of Posidonia oceanica leaf, a molecular tool for marine environment studies. Plant Biosystems 142(2): 213-218.
- Jiang Z, Kumar M, Padula MP, Pernice M, Kahlke T, et al. (2017) Development of an efficient protein extraction method compatible with LC-MS/MS for proteome mapping in two Australian Seagrasses Zostera muelleri and Posidonia australis. Frontiers in Plant Science 8: 1416.
- Piro A, Anagnostopoulou V, Apostolaki ET, Mazzuca S (2022) Fine-tuned method to extract high purified proteins from the seagrass Halophila stipulacea to be used for proteome analyses. Plant Biosystems - An International Journal Dealing with all Aspects of Plant Biology 156(5): 1158-1166.
- Spadafora A, Filadoro D, Mazzuca S, Bracale M, Marsoni M, et al. (2008) 2-DE polypeptide mapping of Posidonia oceanica leaves, a molecular tool for marine environment studies. Plant Biosystems 142(2): 213-218.
- Shevchenko A, Tomas H, Havli J, Olsen JV, Mann M (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nature protocols 1(6): 2856-2860.
- Mi H, Ebert D, Muruganujan A, Mills C, Albou LP, et al. (2020) PANTHER version 16: A revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucl Acids Res 49(D1): D394-D403.
- Laemli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227(5259): 680-685.
- Mazzuca S, Bjork M, Beer S, Felisberto P, Gobert S, et al. (2013) Establishing research strategies, methodologies and technologies to link genomics and proteomics to seagrass productivity, community metabolism, and ecosystem carbon fluxes. Frontiers in Plant Science 4: 38.
- Procaccini G, Beer S, Björk M, Olsen J, Mazzuca S, et al. (2012) Seagrass ecophysiology meets ecological genomics: Are we ready? Mar Ecol 33(4): 522-
- Serra IA, Mazzuca S (2011) Posidonia oceanica: From ecological status to genetic and proteomic resources. In: Pirog RG (Ed.), Seagrass: Ecology, uses and threats. Nova Science Publishers, USA, pp. 71-116.
© 2023 Amalia Piro. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)













.bmp)
.jpg)
.png)
.jpg)














.png)

.png)



.png)






