- Submissions

Full Text
Examines in Marine Biology & Oceanography
Transcriptome Analysis of Immune-Related Gene in Hybrid Yellow Catfish (Tachysurus Vachelli x Tachysurus Fulvidraco) in Reaction to Peptidoglycan Challenge
Tingshuang Pan1,2*, Jun Ling1,2 and Ming Yan1,2
1Fishery Institute of Anhui Academy of Agricultural Sciences, P.R. China
2Key Laboratory of Aquaculture & Stock Enhancement in Anhui Province, P.R. China
*Corresponding author: Tingshuang Pan, Fishery Institute of Anhui Academy of Agricultural Sciences, Key Laboratory of Aquaculture & Stock Enhancement in Anhui Province, Hefei, 230031, P.R. China
Submission: August 23, 2022;Published: December 01, 2022

ISSN 2578-031X Volume5 Issue2
Abstract
Hybrid yellow catfish (Tachysurus vachelli x Tachysurus fulvidraco) is one of the most important freshwater fish in China, intensive culturing has caused increased susceptibility to bacteria, viruses, and parasites. However, little is known about its adaptation mechanisms to pathogen infection. To better understand its immune system and immune related gene under pathogen infection, the transcriptome was analyzed by comparing the data of hybrid yellow catfish spleen stimulated by PGN and PBS. There were 285 Differentially Expressed Genes (DEGs) between PGN treated experimental groups and PBS control groups at 24h, including 213 up regulated and 72 down regulated DEGs. The expression of qRT-PCR from five up regulated DEGs and five down regulated DEGs were consistent with RNA-Seq. In the analysis of GO enrichment, DEGs in the spleens of PGN treated hybrid yellow catfish were significantly enriched in GO items, including biological process, cellular component, and molecular function items. The identified DEGs and enriched GO terms and KEGG pathways were useful for understanding the physical and biochemical performances, and the transcriptome data greatly enriched the genetic information of hybrid yellow catfish.
Keywords: Differentially expressed genes; Genes and genomes; Transcriptome; Ontology
Abbreviations:DEGs: Differentially Expressed Genes; PGN: Peptidoglycan; PBS: Phosphate Buffered Solution; GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes; LPS: Lipopolysaccharide; BP: Biological Process; CC: Cellular Component; MF: Molecular Function
Introduction
Hybrid yellow catfish (Tachysurus fulvidraco♀×Tachysurus vachellii♂) is regarded as a good freshwater species in China for its delicious flesh and high nutrition. In recent years, intensive culturing of hybrid yellow catfish has caused increased susceptibility to bacteria, viruses, and parasites. Pathogen spread and disease outbreak resulted in huge economic loss to hybrid yellow catfish culturing. Knowing more about immune system in hybrid yellow catfish is essential to defense pathogen outbreak and establish effective measures to reduce financial losses. Peptidoglycan (PGN) is a large molecular in cell wall of gram-positive and gram-negative bacteria [1], which has been studied in many fishes as immunostimulan [2-4]. Many defense related genes in fish innate immune system were reported, such as cytokines, chemokines, toll-like receptors, immunoglobulin m, vitellogenin, cellular apoptosis susceptibility, glutathione s-transferase [5]. Proteins of these defense related genes are important in protecting fish against invading viral, bacterial, and fungal. The spleen is an important immune organ and regarded as a primordial secondary lymphoid organ [6], which has been reported to make great contribution to systemic immunity of fish and has the function to remove antigenic substances [7]. RNA-sequencing (RNA-Seq) is a mature technology, which uses high-through sequencing methods to determine all RNA transcripts of the specimen [8]. Transcriptome data can provide a basis to further analyze the immune system in various fishes, such as Pelteobagrus fulvidraco [9-12], Sepia esculenta [13], Paralichthys olivaceus [14], Gymnodiptychus pachycheilus [15], and Salmo trutta [16].
Materials and Methods
Experimental samples
Healthy hybrid yellow catfish (24.60±0.34g) were maintained in 2000L plastic tanks with recirculation system in Fisheries Institute, Anhui Academy of Agricultural Sciences, China. Two weeks later, the fish were divided into an experimental group and control group (twenty fish per group) and transferred into 200L glass tanks. Fish in the experimental group were injected with 100μl baccilus subminis PGN (1mg/ml, 69,554, Sigma) intraperitoneally, and the fish in control group injected with 100ul Phosphate Buffered Solution (PBS). During the experiment, water temperature 26 °C ~ 28 °C, dissolved oxygen 5.0mg/L~6.0mg/L, and pH 7.6 were maintained. Fish were fed with commercial formulated feed. After 24h injection, spleens were collected and immediately frozen in liquid nitrogen, then stored in -80 °C refrigerator. All procedures were carried out according to the Chinese legislation for animal experimentation guidelines.
RNA extraction and library preparation
Total RNA from spleens of hybrid yellow catfish were extracted using Trizol reagent (Sangon, China) according to the manufacturer’s instructions. RNA purity and quantification were evaluated using the Nano Drop 2000 spectrophotometer (Thermo Scientific, USA). RNA integrity was assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies, USA). Then the libraries were constructed using Tru Seq Stranded mRNA LT Sample Prep Kit (Illumina, USA) according to the manufacturer’s instructions. The transcriptome sequencing was conducted by OE Biotech Co., Ltd. (Shanghai, China).
RNA sequencing and transcriptome assembling
The libraries were sequenced on an Illumina HiSeq X Ten platform and 150bp paired-end reads were generated. About 49M raw reads were generated. Raw reads were firstly processed using Trimmomatic [17] and the low quality reads were removed to obtain the clean reads. Then about 48M clean reads were retained for subsequent analyses. The clean reads were aligned to the Tachysurus fulvidraco reference genome (https://www.ncbi.nlm. nih.gov/genome/?term=yellow+catfish) using HISAT2 package to obtain annotation information [18].
Identification of differentially expressed genes
Each gene was obtained by HTSeq-count [19]. Differentially expressed genes (DEGs) were analyzed using DESeq (2012) R package [20]. The threshold for DEGs was set to q value <0.05 and |log2foldchange|≥1. All DEGs were mapped to the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) [21] databases to determine their potential function and metabolic pathway.
Quantitative real-time PCR (qRT-PCR) verification
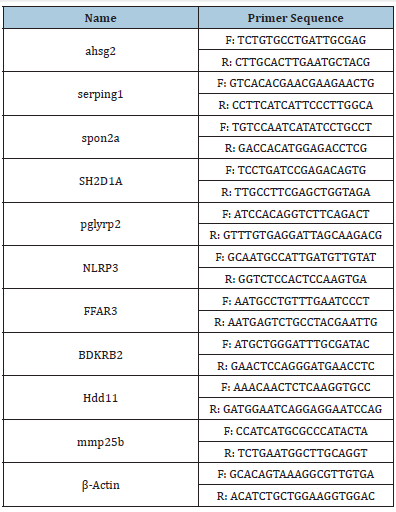
To validate the results of RNA-Seq, ten DEGs were selected to analysis qRT-PCR by SYBR Premix Ex Taq kit (Invitrogen). RNA samples used for qRT-PCR were the same as the RNA used for transcriptome. Primers of qRT-PCR were listed in (Table 1). All reactions were performed using technical triplicates. β-Actin was selected as the reference gene. Relative expression was calculated by the 2-ΔΔCt method [22].
Table 1:Sequences of primers used for qRT-PCR.
Results
Transcriptome sequencing and annotation of the unigene
Transcriptomes of the spleens from hybrid yellow fish treated by PBS or PGN were sequenced. After removing low quality sequences, number of clean reads in these six libraries ranged from 47424226 to 50845358, and the clean bases ranged from 7.04G to 7.54G. Q30 values of the sequenced libraries ranged from 93.05% to 94.98%, whereas the GC content of the libraries ranged from 47.22% to 47.40% (Table 2). These results confirmed that the sequenced data and transcriptome were reliable. In the transcriptome data for CS1, CS2, CS3 (PBS group), and ES1, ES2, ES3 (PGN group), the reads and percentage mapped to the yellow catfish genome is 36809302, 37225718, 39265044, 37309076, 37984493, 37472668, and 77.62%, 77.30%, 77.22%, 76.91%, 76.67%, 76.84%, respectively (Table 3). The raw reads of RNA-Seq were deposited in the Sequence Read Archive database of NCBI with the accession numbers of PRJNA833861.
Table 2:Overview of spleen transcriptome sequencing data and quality filtering.

Table 3:Transcriptome reads, and percentage mapped to the yellow catfish genome.

Identification and analysis of DEGs
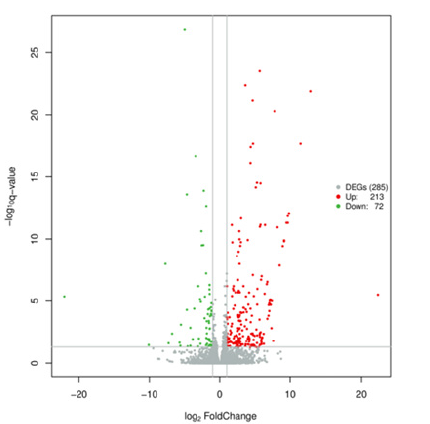
The DEGs between PGN treated experiment groups and PBS control groups were identified on the basis of q value <0.05 and |log2foldchange|≥1. There were 285 DEGs between PGN treated experimental groups and PBS control groups at 24h, including 213 up regulated and 72 down regulated DEGs (Figure 1). In the volcano plot, significantly up regulated unigenes represented by red dots, significantly down regulated unigenes represented by green dots, and no significantly expressed unigenes represented by grey dots. The levels of magnitude changes in up regulated DEGs are lower than that of down regulated DEGs. When yellow catfish challenged with Lipopolysaccharide (LPS), 27 immune-related genes and 7 transcriptional genes were identified [23]. 765 up regulated and 1757 down regulated genes were obtained from hepatopancreas, when Procambarus clarkii was challenged with PGN [24]. 466 up regulated genes and 834 down regulated genes were obtained from Sesarmops sinensis hepatopancreas when challenged by PGN [25]. Transcription levels of G-CSFR, BPI/LBP and HSP70-4 were increased from the kidney of Cyprinus carpio by PGN stimulation [3].
Figure 1:Volcano plot of Differentially Expressed Genes (DEGs) from PGN-treated and PBS control groups in hybrid yellow catfish. The x-axis represents the fold change between the PGN and PBS groups; the y-axis indicates the significance of differential expression. Grey dots represented no significantly expressed unigenes, while red and green dots represent up- and down- regulated unigenes, respectively (q value <0.05 and |log2foldchange|≥1).
Functional annotation by GO and KEGG analysis
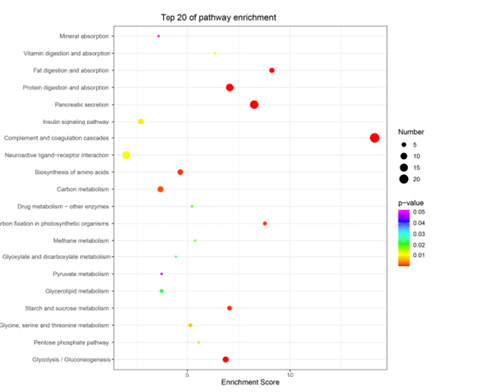
In the analysis of GO enrichment, DEGs in the spleens of PGN treated hybrid yellow catfish were significantly enriched in 134 GO items, including 89 Biological Process (BP), 16 Cellular Component (CC), and 29 Molecular Function (MF) items. The significantly enriched five top BP items were “negative regulation of endopeptidase activity”, “proteolysis”, “complement activation, classical pathway”, “complement activation, alternative pathway”, and “digestion”. The significantly enriched five top CC items were “extracellular space”, “extracellular region”, “blood microparticle”, “membrane attack complex”, and “cortical granule”. The significantly enriched five top CC items were “serine-type endopeptidase activity”, “serine-type endopeptidase inhibitor activity”, “endopeptidase inhibitor activity”, “cysteine-type endopeptidase inhibitor activity”, and “complement component C3b binding” (Figure 2). These results indicate that substantial changes emerged in energy metabolism and immune defenses in spleens under PGN stress. The KEGG database was used to investigate biochemical pathways of DEGs. The top 20 KEGG pathways included mineral absorption, vitamin digestion and absorption, fatty digestion and absorption, protein digestion and absorption, pancreatic secretion, insulin signaling pathway, complement and coagulation cascades (Figure 3).
Figure 2:Gene Ontology (GO) TOP 30 terms enriched by Differentially Expressed Genes (DEGs). The results are summarized in three main categories: Biological process, cellular component, and molecular function. The x-axis indicates the second term of GO and the y-axis indicates gene percentage.
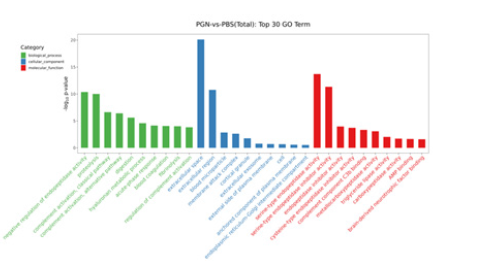
Figure 3:Top 20 significantly enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways of DEGs. Color and gene ratio indicate the P-value and the ratio of genes with each pathway, respectively. The x-axis indicates the enrichment score of KEGG and the y-axis indicates the name of the top 20 pathways.
qRT-PCR analysis
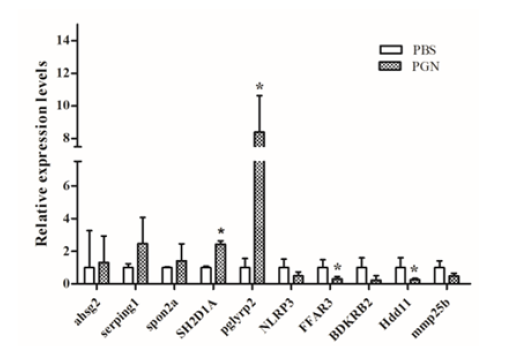
Five upregulated DEGs (ahsg2, serping1, spon2a, SH2D1A, pglyrp2) and five downregulated DEGs (NLRP3, FFAR3, BDKRB2, Hdd11, mmp25b) were chosen for qRT-PCR analysis. The results showed the expression of qRT-PCR were consistent with that of RNA-Seq (Figure 4). The results showed that serping1, SH2D1A, pglyrp2, were upregulated at least two times, and all the downregulated DEGs were downregulated at least two times of the control. Pglyrp2, BDKRB2, and Hdd11 were particularly responsive to the PGN stimulation. In our study, pglyrp2 was upregulated eight times in hybrid yellow catfish in response to PGN, Whereas BDKRB2 and Hdd11 were downregulated four times when compared to the control. These immune related genes may be involved in innate immune defense against PGN in T. fulvidraco.
Figure 4:Five upregulated and five downregulated immune related genes treated by PGN stimulation relative to PBS were selected to validate RNA-seq by qRT-PCR.
Discussion
In our study, transcriptome profiles of hybrid yellow catfish was used to invest immune mechanism when the fish challenged with PGN. Tanscriptome sequencing results showed that immune related DEGs occupied large portion of all the DEGs. Pglyrp2 was an inducible and constitutive acute-phase protein [26] and had antimicrobial and peptidoglycan-lytic amidase activity in fish [27,28]. In our study, the expression of pglyrp2 was up-regulated when the hybrid yellow catfish was challenged with PGN, suggesting that pglyrp2 has important role in recognizing bacteria and initiating downstream antimicrobial pathway. BDKRB2 is a component of kallikrein-kinin system, which participates in physiological and pathological processes [29]. Current study showed that BDKRB2 was downregulated when challenged by PGN. Previously study showed that BDKRB2 were expressed relatively higher at the time of pre-fertilization and pre-hatching in the ovarian of rockfish [29]. Bdkrb2 was also related to cerebrovascular dysfunction with the development of rat’s Alzheimer’s disease and expressed lower at 20 day and 18 month [30]. HDD11 is an innate immune protein, which showed transcriptional suppression in our study. HDD11 transcripts significantly lower expressed (~25 fold) in sea slice parasitized group compared with the non-infected group , suggesting that sea slice can exert immunosuppressive in fins of salmon [31].
Conclusion
In conclusion, we investigated the transcriptome of hybrid yellow catfish immune response upon PGN injection. A total of 285 DEGs between PGN treated experimental groups and PBS control groups at 24h, including 213 up regulated and 72 down regulated DEGs. These DEGs contribute to anti-bacterial responses after yellow catfish challenged with PGN. Results of our study could be benefit for better understanding the defense mechanisms and innate immune system of hybrid yellow catfish by the challenge of PGN, and valuable information for further studying on fish innate immune system.
References
- Wolf AJ, Underhill DM (2018) Peptidoglycan recognition by the innate immune system. Nature Reviews Immunology 18(4): 243-254.
- Kondo H, Kikumoto T, Yoshii K, Murase N, Yamada H, et al. (2021) Effects of peptidoglycan and polyinosinic: Polycytidylic acid on the recombinant subunit vaccine efficacy against edwardsiella tarda in Japanese flounder paralichthys olivaceus. Fish Pathology 56(3): 149-155.
- Kono T, Ponpornpisit A, Sakai M (2004) The analysis of expressed genes in head kidney of common carp Cyprinus carpio stimulated with peptidoglycan. Aquaculture 235(1): 37-52.
- Su Y (2011) Isolation and identification of pelteobagrin, a novel antimicrobial peptide from the skin mucus of yellow catfish (pelteobagrus fulvidraco). Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology 158(2): 149-154.
- Abo-Al-Ela HG (2018) An introduction to selected innate immune-relevant genes in fish. Applied Ecology and Environmental Research 16(2): 955-976.
- Flajnik MF (2018) A cold-blooded view of adaptive immunity. Nature Reviews Immunology 18(17): 438-453.
- Mebius RE, Kraal G (2005) Structure and function of the spleen. Nature Reviews Immunology 5: 606-616.
- Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y (2008) RNA-seq: An assessment of technical reproducibility and comparison with gene expression arrays. Genome Research 18(9): 1509-1517.
- Chen X, Mei J, Wu J, Jing J, Ma W, et al. (2015) A comprehensive transcriptome provides candidate genes for sex determination/differentiation and SSR/SNP markers in yellow catfish. Marine Biotechnology 17(2): 190-198.
- Liu Y, Xin ZZ, Zhang DZ, Wang ZF, Zhu XY, et al. (2017) Transcriptome analysis of yellow catfish (Pelteobagrus fulvidraco) liver challenged with polyriboinosinic polyribocytidylic acid (poly I: C). Fish & Shellfish Immunology 68: 395-403.
- Tao YF, Qiang J, Dagoudo M, Zhu HJ, Bao JW, et al. (2021) Transcriptome profiling reveals differential expression of immune-related genes in gills of hybrid yellow catfish (tachysurus fulvidraco♀×pseudobagrus vachellii♂) under hypoxic stress: Potential NLR-mediated immune response. Fish & Shellfish Immunology 119: 409-419.
- Zhang J, Ma W, Song X, Lin Q, Gui JF, et al. (2014) Characterization and development of EST-SSR markers derived from transcriptome of yellow catfish. Molecules 19(10): 16402-16415.
- Bian L, Liu C, Chen S, Zhao F, Ge J (2018) Transcriptome analysis of gene expression patterns during embryonic development in golden cuttlefish (Sepia esculenta). Genes Genomics 40(3): 253-263.
- Kim WJ, Lee K, Lee D, Kim HC, Nam BH, et al. (2021) Transcriptome profiling of olive flounder responses under acute and chronic heat stress. Genes & Genomics 43(2): 151-159.
- Yang L, Wang Y, Zhang Z, He S (2014) Comprehensive transcriptome analysis reveals accelerated genic evolution in a tibet fish, gymnodiptychus pachycheilus. Genome Biology and Evolution 7(1): 251-261.
- Carruthers M, Yurchenko AA, Augley JJ, Adams CE, Herzyk P, et al. (2018) De novo transcriptome assembly, annotation and comparison of four ecological and evolutionary model salmonid fish species. BMC Genomics 19(1): 1-17.
- Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30(15): 2114-2120.
- Kim D, Langmead B, Salzberg SL (2015) HISAT: A fast spliced aligner with low memory requirements. Nature Methods 12(4): 357-360.
- Anders S, Pyl PT, Huber W (2015) HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 31(2): 166-169.
- Anders S, Huber W (2012) Differential expression of RNA-Seq data at the gene level-the DESeq package. European Molecular Biology Laboratory (EMBL), pp. 1-24.
- Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, et al. (2007) KEGG for linking genomes to life and the environment. Nucleic Acids Research 36 :480-484.
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-ΔΔC(T) method. Methods 25(4): 402-408.
- Liu QN, Xin ZZ, Chai XY, Jiang SH, Li CF, et al. (2016) Characterization of immune-related genes in the yellow catfish Pelteobagrus fulvidraco in response to LPS challenge. Fish & Shellfish Immunology 56: 248-254.
- Chu XH, Yang TT, Liu Y, Hong L, Jiao T, et al. (2019) Transcriptome analysis of differential expressed genes in hepatopancreas of Procambarus clarkii challenged with peptidoglycan. Fish & Shellfish Immunology 86: 311-318.
- Li YT, Tang BP, Zhang SP, Tang YY, Wang G, et al. (2021) Transcriptome analysis of immune-related genes in Sesarmops sinensis hepatopancreas in reaction to peptidoglycan challenge. Genomics 113(3): 946-954.
- Mao Y, Wang J, Zhang Z, Ding S, Su Y (2010) Cloning, mRNA expression, and recombinant expression of peptidoglycan recognition protein II gene from large yellow croaker (Pseudosciaena crocea). Molecular Biology Reports 37(8): 3897-3908.
- Choi KM, Joo MS, Cho DH, Bae JS, Jung JM, et al. (2019) Characterization of gene expression profiles and functional analysis of peptidoglycan recognition protein 2 from rock bream (Oplegnathus fasciatus). Fish & Shellfish Immunology 84: 1068-1074.
- Li X, Wang S, Qi J, Echtenkamp SF, Chatterjee R, et al. (2007) Zebrafish peptidoglycan recognition proteins are bactericidal amidases essential for defense against bacterial infections. Immunity 27(3): 518-529.
- Niu J, Song W, Li R, Yu H, Guan J, et al. (2021) The Bdkrb2 gene family provides a novel view of viviparity adaptation in Sebastes schlegelii. BMC Ecology and Evolution 21: 1-11.
- Stefanova NA, Maksimova KY, Rudnitskaya EA, Muraleva NA, Kolosova NG (2018) Association of cerebrovascular dysfunction with the development of Alzheimer’s disease-like pathology in OXYS rats. BMC Genomics 19(3): 51-63.
- Umasuthan N, Xue X, Caballero-Solares A, Kumar S, Westcott JD, et al. (2020) Transcriptomic profiling in fins of Atlantic salmon parasitized with sea lice: Evidence for an early imbalance between chalimus-induced immunomodulation and the host’s defense response. International Journal of Molecular Sciences 21(7): 2417.
© 2022 Tingshuang Pan. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






