- Submissions

Full Text
Developments in Clinical & Medical Pathology
ESKAPE Pathogens: Infection, Mode of Resistance and Its Cure
Umesha S* and Suguna M
Department of Biotechnology, University of Mysore, Manasagangotri, India
*Corresponding author: Umesha S, Department of Biotechnology, University of Mysore, Manasagangotri, India
Submission: August 08, 2022;Published: October 19, 2022

ISSN:2690-9731 Volume2 Issue2
Abstract
Generally infectious diseases are caused by organic agents such as viruses, bacteria, parasites, fungi protozoans and other microbes, which can spread directly through contact with an infected person or animal. The means of spread is through consumption of contaminated food or water, contacting contaminated surroundings such as infected animal droppings or inhaling contaminated air, through biting of insects like mosquitoes or ticks [vectors] carrying the agents. Studies have proven, that demographic and environmental factors such as population growth, increased urbanization and evolution of disease-carrying insects and animals have contributed greatly to the spread of these deadly diseases. In this scenario, major challenge for the medical and scientific world is to find solutions via discovering new synthetic or natural agents which can treat these infectious diseases. Many strategies and techniques are being employed to discover these cryptic agents from decades and as the new diseases emerge, the idea of discovering more and more of these agents is renewed t suit the new challenge.
Keywords: ESKAPE pathogens; Mechanism of drug resistance; Antibiotics; Plant secondary metabolites
Introduction
Infectious diseases are human illnesses caused by viruses, bacteria, parasites, fungi, and other microbes [1]. They can be spread by direct contact with an infected person or animal, through ingestion of contaminated food or water, by insects like mosquitoes or ticks [disease vectors], or by contact with contaminated surroundings such as through touching animal droppings or breathing in contaminated air [2]. Environmental factors such as growth of population, urbanization, alteration of habitats of disease-carrying biosystems have contributed to the spread of infectious diseases [2]. CDC has listed out these emerging issues of antibiotic resistance as “biggest threats” as these bugs are rapidly evolving and finding new techniques to develop resistance against antibiotics [3]. The race between pathogens which are developing resistance and world of medical science to discover new strategy or drug to combat these pathogens has become very critical, as these bugs are out racing with speed the pace of discovering novel drugs against them [1].
ESKAPE pathogens
According to CDC, an estimate of over 2 million illnesses and nearly 23,000 deaths per year were caused by antibiotic resistant ESKAPE pathogens [1]. To focus and alert the scientific world, CDC has coined the term ‘ESKAPE pathogens’ to represent these highly contagious and deadly bacteria with ability of multi-drug resistance. Off late, the deadly infections caused by these bacteria are more prevalent in hospital settings, called Hospital-Acquired Infections [HAIs] [4]. These ESKAPE pathogen are grouped into as following
E for Enterococcus faecium
Enterococcus faecium is a Gram-positive bacterium that is famous to cause wound infections, urinary tract infections, endocarditis, and nosocomial bacteraemia [5]. The word nosocomial refers to infections which are acquired in hospitals and the word bacteraemia means, the critical clinical condition where, presence of bacteria can be traced in blood, which is often fatal [6]. According to epidemiological report of October 1990, E. faecium strain having antibiotic resistance to glycopeptides, penicillin, and aminoglycosides was isolated from a patient in an ICU [intensive care unit] [5]. Following this report, these pathogenic strains of E. faecium with multiple drug resistance capacity were isolated from eight other patients in their blood, urine, or surgical wound specimens pointing towards the rapid rate of spreading of these infectious strains and their highly contagious nature [6]. Approximately 110,000 cases per year are reported to be caused by Urinary Tract Infections [UTI], which are the most prevalent disease caused by these multi drug resistant E. faecium [6]. Many of the patients acquire these infections via nosocomial, they are those who had become immunocompromised due to their prolonged hospital stay, exposure, pre-treatments etc and makes them more prone and vulnerable to these ESKAPE pathogens [5]. E. faecium have now become resistant against many new generation and strong antibiotics such as Penicillin, Gentamicin, Tetracycline, Erythromycin etc. Even to Vancomycin, which is now a last resort treatment available for infections caused by Gram-positive bacteria [6].
S for Staphylococcus aureus
Staphylococcus aureus belongs to Gram-positive bacterium category and is normally found in nose and skin of healthy people [4]. When tested in human subjects, one in three were found to be carrying S. aureus in their nose without any illness, which is known as asymptomatic colonization S. aureus [7]. This bug is known to cause skin and soft tissue infections in majority of the population which either hospital or community acquired [3]. At its severity of infection, S. aureus is known to causes bacteraemia which is highly difficult to treat and fatal. Through the techniques of gene transfer, S. aureus possesses and acquires resistance to most recent antibiotics, best example is Methicillin, which has created a unique superbug called Methicillin Resistant S. aureus [MRSA] [3]. Misuse of antibiotic treatment over the course of decades has led to the emergence of this superbug MRSA. Based on reports issued by CDC, this superbug is found to cause infections, at around 80,000 cases and 11,000 deaths each year [7].
K for Klebsiella pneumoniae
Klebsiella pneumoniae is a nonmotile, Gram-negative bacterium exist as normal flora of the mouth, skin and intestines but in low numbers [8]. However, K. pneumoniae is known cause lower respiratory organ like lung infections [2]. K. pneumoniae causes infections such as inflammation, necrosis, and haemorrhage in lung tissue and can also result in UTIs [8]. This bug mainly infects, patients on ventilators, catheters or with surgical wounds and these infections are majorly hospital acquired [8]. K. pneumoniae is more prevalent in ICUs and it is raising alarm since it is found to have gained resistance against carbapenems [2]. Carbapenems are last resort antibiotics being used to treat patients suffering from infection acquired by these multidrug resistant bugs at hospitals [8]. Unfortunately, Carbapenem Resistant Klebsiella pneumoniae [CRKP] have emerged which are found to be resistant to almost all available antibiotics and the infections caused by these super bugs are associated with high rates of mortality [2].
A for Acinetobacter baumannii
Acinetobacter baumannii is a Gram-negative bacterium that are found typically in respiratory secretions, wounds, and urine of hospitalized patients. It can also be found in solutions which are used for irrigation of wounds and intravenous solutions in the hospital setting [1].
P for Pseudomonas aeruginosa
Pseudomonas aeruginosa belongs to Gram-negative class of bacteria. These are opportunistic pathogens having mortality rate of about 60%. This bacterium is most found in hospitalized patients. P. aeruginosa has now found to have developed resistance to antibiotics rapidly such as Ciprofloxacin and Levofloxacin [9]. This pathogen has the capability of forming a biofilm on surfaces, through which it creates a “shield” making it extremely strong for any antibiotic to destroy, specifically in cystic fibrosis patients [1]. So, this MDR strain of P. aeruginosa especially found in wound infections has become major concern globally because of its difficulty in treatment. Infections caused by these multi drug resistant P. aeruginosa has become unarguably a humongous challenge to the modern medicine field [1].
E for Enterobacter species
Enterobacter species are a genus that comprises Gram-negative bacteria which mostly infect lower gastrointestinal tract like kidney and respiratory tracts region such as lungs [10]. These genus pathogens are found to be resistant to many generations of Penicillin and Cephalosporins [11]. Beta-lactam are the bestknown antibiotics used for the treatment of the diseases caused by the pathogens, but new reports have alerted on the emergence of new β-lactamase-mediated resistance in multidrug-resistant Enterobacter at alarming rate of spreading, which is a major concern in treating HAIs now a days [12].
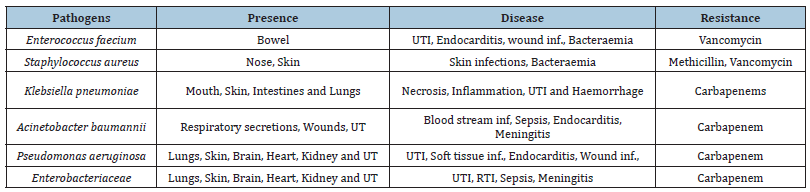
A major challenge is therefore to find ways to elicit the new class of yet unknown antibiotics with novel mode of action. Such strategies have been in use in the scientific field for many decades, but the discovery of these new and cryptic antibiotics has led to the path of new infections and mechanism of drug resistance by pathogens [2]. Below table list out all ESKAPE pathogens, their presence in the body, diseases caused by them and the most recent or last resort antibiotics to which they are found to acquire resistance (Table 1).
Table 1: List of ESCAPE pathogens.
Resistance Mechanisms of ESKAPE Pathogens
There are many mechanisms acquired by these ESKAPE pathogens such as antimicrobial resistance genes, carried on the bacterial chromosome, plasmid, or transposons and these helps to develop resistance towards the antibiotics [13]. Different mechanisms of drug resistance adopted by different pathogens are classified into broad categories, such as inactivation/alteration of drug, modification of drug binding sites/targets modification, comprising cell permeability which causes reduced drug accumulation inside the cell and biofilm formation etc [14].
Drug inactivation or alteration
Pathogen have picked up easy drug resistance mechanism through enzyme production such as 𝛽-lactamases, amino glycosidases, chloramphenicol acetyltransferases etc. these enzymes modify and inactivate the antibiotics irreversibly and make them inactive [15]. 𝛽-lactamases are one the well-studied enzyme and are highly prevalent [16]. 𝛽- lactamases causes hydrolyzation of 𝛽-lactam ring of 𝛽-lactams; thus, the antibiotics which fall under this group of 𝛽-lactams such as penicillin, cephalosporins, monobactams, and carbapenems are inhibited through this strategy [17].
Modification of drug binding sites
In this mechanism, the pathogens modify their target sites so that antibacterial agents lose the ability to recognise and binding [17]. The best example to know about this mechanism are the Penicillin- Binding Proteins [PBP]. The genes code for the mutation of these PBPs which typically anchored on the cytoplasmic membrane of cell wall of bacteria, and they function in assembly to regulate different stages of cell wall building etc. These are specialized and strong in their function, for instance, the expression of PBP2a in S. aureus, which is a unique and dominant enzyme in MRSA compared to PBPs present in sensitive S. aureus [18]. PBP2a expressed in MRSA have lower affinity towards almost all 𝛽-lactams and they also act as a substitute for the other PBPs, [PBPs 1-4] thus assisting MRSA to survive easily even in the presence of high concentrations of 𝛽-lactam drugs including methicillin. Another example for this mechanism is gene mutation caused in peptidoglycan precursors. In methicillin-resistant Gram-positive organisms, bacterial cell wall synthesis can be inhibited by glycopeptides, targeting acyl- D-alanyl-D-alanine [acyl D-Ala-D-Ala] residues of peptidoglycan precursors [19]. This is achieved by mutating the crosslink target of peptidoglycan to [D-Ala-D-Ala to D-Ala-D-Lac or D-Ala-D-Ser], which is carried out by yet another complex gene cluster such as [Van-A, Van-B, Van-D, Van-C, Van-E, and Van-G]. By applying these strategies, E. faecium and E. faecalis enhance their resistance to glycopeptides against the much more strong and recent class antibiotics such as vancomycin and teicoplanin [19].
Reduced intracellular drug accumulation
Cell membrane plays a major role in maintaining the balance of uptake and elimination which is the key for the bacteria to maintain its good health. Cell membranes not only regulate fluids intake and elimination but also determines the in and out flow of antibiotics which defines the susceptibility of bacteria to a particular drug [15]. Thus, adopting a strategy where if bacteria reduce the amount of antibiotic permeability and thereby disables the antibiotics to pass through the cell membrane will prove to be useful for the bacteria to develop antibiotic resistance [17]. This mechanism is made possible by bacteria through expressing diminished protein channels on their outer membrane to avoid drug entry or expressing efflux pumps to pump out and decrease the amount of drug already accumulated within the cells [13].
Porin loss
Another similar strategy picked by Gram-negative bacteria are the proteins called porins. These are transport proteins which are found in the outer membrane pathogens whose function is to allow the passage of hydrophilic substances, including antibiotics through the channels formed by them [18]. A reduction in the number of porin proteins ill restrict the flow of antibiotics, this same strategy is picked up by P. aeruginosa porin protein OprD which resulted in impaired drug inflow into the cell, thus pathogen avoids itself to exposure of deadly antibiotics such as imipenem and survives. Another Gram-negative bacterium, such as A. baumannii, sheds its Outer Membrane Protein [OMP] and becomes resistive to imipenem and meropenem antibiotics. In MDR strains of K. pneumoniae, the outer membrane proteins such as OmpK35and OmpK36 and by producing enzymes known as AmpC 𝛽-lactamase and newgeneration carbapenems A, aid in developing resistances and exhibits reduced susceptibility to 𝛽-lactams such as cephalosporins and carbapenems [19].
Efflux pumps
Some pathogens have developed resistance mechanism through efflux pumps. These efflux pumps are the exporters which help to remove antibiotics from inter membrane space or intracellular compartment in some Gram- negative bacteria [16]. The rate of expelling the antibiotics is high with these efflux pumps, that the drug concentrate never reaches the lethal or effective dose inside the bacteria to elicit its antimicrobial effect. Most of these efflux pumps expressed on the cell membrane surface are designed to expel a wide range of antibiotics, thus pathogens owning these efflux pumps usually show multidrug resistance [15]. Till date, the efflux pumps identified, belong to five super families such as ATPBinding Cassettes [ABC], Resistance-Nodulation-Division [RND], major facilitator super family, small multidrug resistance family, and multidrug and toxic compound extrusion family [18].
Polyselective efflux pumps which belong to RND family are most common type of efflux pump present in Gram-negative bacteria, that plays a major role in expressing Multidrug Resistance [MDR] bacterial phenotype [17]. These efflux pump helps to get rid of a variety of antibiotics which can be structurally different from each other, such as dyes and salts, detergents and biocides that are frequently used in medical field and treatments [14]. The best known of these RND efflux pumps are AcrAB-TolC and MexAB-OprM and are usually chromosomally encoded [18]. These two efflux pumps aids pathogen to survive most toxic agents thus becoming essential part of bacteria [16].
For example, bacterium P. aeruginosa possess four potent RND type multidrug resistance efflux pumps called Mex, that can expel out antibiotics and other toxic compounds from periplasm and cytoplasm [13]. Two efflux pumps, namely MexAB-OprM and MexCD-OprJ, which belong to this family help to eliminate three main classes of antibiotics, such as carbapenems, fluoroquinolones, and aminoglycosides [18]. Now a days, it is reported that, in many clinical isolates of P. aeruginosa, over expression of efllux pumps MexCD-OprJ and MexEF-OprN is observed which indicates the emergence of more and mor MDR strains [17]. Also, increase in the prevalence of these kind of efflux pumps and its overproduction is reported in other pathogens such as Enterobacter aerogenes and K. pneumoniae clinical isolates [13]. Similarly, the presence and overexpression of AdeABC efflux pumps which belong to the family RND and low porin expression, marks the emergence of MRD strains of A. baumannii [15]. AdeABC efflux pumps are highly efficient in expelling broad range of antibiotics, such as fluoroquinolones, 𝛽-lactams, tetracyclines, macrolides, chloramphenicol, and aminoglycosides [18].
Existing antibiotics and their mode of action
The term antibiotic originated from the word, ‘antibiosis’ which means ‘against life’. Earlier, antibiotics were those organic compounds produced by one microorganism on account of their defence and are toxic to other microorganisms [8]. An antibiotic can be defined as a substance, produced by one microorganism, or of biological origin at low concentrations which can inhibit the growth of, or are lethal to other microorganisms [2]. However, this definition doesn’t hold good enough now and it is modified in modern times, to also include all those antimicrobials that are also produced partly or wholly through synthetic means along with naturally produced antibiotics.
Some antibiotics have the capability to completely kill other invaders, and some are only able to inhibit their growth [12]. Although antibiotic generally refers to antibacterial, but it is broadly classified as antibacterial, antifungals and antivirals to represent the group of invaders/microorganisms they antagonize [19]. There are many ways to classify antibiotics. But the best classification strategy is based on their molecular structures, mechanism of action and spectrum of activity. Some of the common antibiotics are betalactams, macrolides, tetracyclines, quinolones, aminoglycosides, sulphonamides, glycopeptides and oxazolidinones [2].
Beta-lactams
This antibiotic class are marked by the presence of a 3-carbon and 1-nitrogen ring that is highly reactive [19]. The ring structure present is very essential in interfering with bacterial proteins, which essential for synthesis of cell wall, and in the process with these ring structure the beta-lactams end up becoming lethal to the bacteria or they inhibit bacterial growth [16]. For instance, beta-lactam antibiotics bind themselves to these Penicillin-Binding Protein [PBP] enzymes. these bacterial enzymes called PBP are those responsible for cross linking of units of peptides during peptidoglycan synthesis [20]. So, this binding of beta-lactams to PBP enzymes interfere with the peptidoglycan synthesis, causing either lysis or cell death [19]. Most prominent and effective beta-lactam are penicillin, cephalosporins, monobactams and carbapenems etc [20].
Macrolides
In 1952, j. M. Mcguire, discovered and isolated antibiotics belonging to this class of called macrolides, as a metabolic by product from a soil inhabiting fungus called Saccharopolyspora eryhraea [16]. The mode of action in either killing or inhibiting the bacterial growth of this class of antibiotics is inhibition of bacterial protein synthesis. This inhibition is achieved by binding to the ribosome, and in the process, to prevent the addition of amino acid to the growing polypeptide chains during bacterial protein synthesis [21-26]. Macrolides are generally broad spectrum, killing or inhibiting both gram-positive and gram-negative pathogens. Commonly known macrolides are erythromycin, azithromycin and clarithromycin [27].
Tetracyclines
Benjamin Duggar discovered this class of antibiotics called tetracycline in 1945 from a soil bacterium of the genus streptoyces [28]. Even this class of antibiotics inhibit the bacterial protein synthesis similar to macrolides. Antibiotics belonging to this class are classified into different generations based on their method of synthesis [16]. Those which are obtained from biosynthesis are said to belong first generation antibiotics such as tetracycline, chlortetracycline, oxytetracycline and demeclocycline [15]. Other members of this class of tetracyclines such as doxycycline, lymecycline, meclocycline, methacycline, minocycline, and rolitetracycline etc are considered second generation are they are derivatives of semi-synthesis. Those which came from total synthesis such as tigecycline are considered as third generation [28].
Quinolones
Group of scientists were involving in the search of antimalarial drugs and accidentally discovered this class antibiotics [19]. It was first discovered as nalidixic acid, as an impurity during the development of quinine [24]. The mechanism of action of quinolones is that they interfere with DNA replication and transcription in bacteria [26]. There are two major groups of antibiotics have been developed from this basic molecule: such as quinolones and naphthyridones. Best examples are Cinoxacin, norfloxacin, ofloxacin, ciprofloxacin, temafloxacin, sparfloxacin, nalidixic acid, enoxacin etc [19]. Structurally these antibiotics consist of two rings, which help them to interfere with replication and transcription. But in recent quinolones, there is one ring structure attached enabling them to extend their spectrum of antimicrobial activity against some anaerobic bacteria that were resistant to two ring quinolones till now [26]. Slight modifications and improvisation of the basic structure of this class of antibiotics are proven to have increase their spectrum of activity and potency and improve bioavailability [24].
Aminoglycosides
This class was first discovered in 1943 obtained from soil bacteria Actinomycetes and the first one to be discovered among its members was streptomycin [14]. Streptomycin has been used extensively to treat tuberculosis caused by M. tuberculosis [14]. In this class, the basic structure usually contains 3-amino sugars connected by glycosidic bonds [16]. Aminoglycosides show broad spectrum of antibacterial activity by killing both gram positive and negative bacteria [22]. Their main mode of action is through inhibiting the protein synthesis by binding to ribosomal subunits during bacterial protein synthesis. In spite of its strong potency towards killing pathogen, it is also found to be toxic to humans, so there is constant search for new class of aminoglycosides which is effective against pathogen but safe to humans [14]. There are many such aminoglycosides were discovered namely gentamicin, neomycin, tobramycin and amikacin [25]. Gentamicin is less toxic to human but are found to be very effective against gram-negative pathogens, hence it is used to treat infections caused by bacteria such as E. coli, P. aeruginosa, Shigella and Salmonella species [15].
Sulphonamides
Sulphonamides are broad spectrum antibiotics inhibit both Gram-positive and Gram-negative bacteria such as E. coli, K. pnemoniae, C. trachomatis Salmonella, Shigella and Enterobacter species, and some Protozoans, and are widely used in the treatment of various infections caused by these bacteria such as septicemia, meningococcal meningitis, bacillary dysentery and urinary tract infections [26]. It is also reported that sulphonamides are also able to impede cancerous cell agents [14]. The mode of action of this class is mainly inhibiting bacterial enzymes and thus, sulphonamides are bacteriostatic rather than bactericidal [26].
Oxazolidinones
This class of antibiotics are relatively a new class which are obtained synthetically and are approved only recently to treat infections. The first member of its member called linezolid was synthesized and approved for medical use only recently in the year 2000 [24]. Although the mechanism of action of this class of antibiotics is not fully understood, but on the surface, it is found to interfere with protein synthesis [25]. Oxazolidinones elicit their action by binding to the P site of 50S ribosomal subunit and there by inhibiting protein synthesis [24]. They have a very broad spectrum of activity against Gram-positive bacteria and also some the MDR strains such as methicillin-resistant staphylococcus species and vancomycin-resistant staphylococcus species, vancomycinresistant enterococcus species, penicillin-resistant pneumococcus species and some of the anaerobes [24].
Glycopeptides
Glycopeptides abbreviated as GPAs were obtained as natural products initially, but off late many semi-synthetic glycopeptides were obtained which showed improved activity and good pharmacokinetic aspects [8]. Naturally obtained glycopeptides contains cyclic peptide of 7 amino acids, bound to 2 sugars, hence the name glycopeptides [10]. The mode of action of this class is through binding to its target via the formation of 5 hydrogen bonds with the peptide backbone. In some other cases such as oritavancin, an additional chlorine and/or sugar is/are attached to the backbone, during the synthesis [8]. These attachments are known to bind more efficiently to target in bacteria and thereby increasing the potency of glycopeptides [2].
Potent antimicrobial agents derived from plants
Since very long, Plants have become the most important therapeutic sources available to us [20]. Natural products, such as plants extract, either as pure compounds or as standardized extracts, provide unlimited opportunities for new drug discoveries because of the unmatched availability of chemical diversity [21]. According to the World Health Organization [WHO], more than 80% of the world’s population relies on traditional medicine for their primary healthcare needs [20]. From the phytochemical and pharmacological viewpoint, only a relative less percentage of plant species are studies [21]. Plant kingdom still is treasure for novel molecule which can combat deadly microorganisms
Thymol, a plant-derived antimicrobial agent, caused rapid efflux of intracellular constituents of P. gingivalis, S. artemidis and S. sobrinus [23]. Studies suggest that membrane perforation is a principal mode of action of this substance. The thymol-induced decline of intracellular ATP in S. sobrinus and it appears to be entirely attributable to leakage, whereas in P. gingivalis thymol may also inhibit ATP generating pathways [23]. Leaf extract of the of E. duttonii, a traditional Australian medicinal plant previously shown to have potent bactericidal activity against gram-positive bacteria [22]. The extract compromised the integrity of the cytoplasmic membrane of S. aureus, leading to increased membrane permeability [indicated by uptake of PI] and a decreased ability to exclude NaCl. The bactericidal action of the E. duttonii extract was concluded to be due to its membrane-active properties [23].
Antibacterial effects of three terpene alcohols on S. aureus, revealed that terpene alcohols, namely, farnesol, nerolidol and plaunotol might act on cell membranes [23]. The antibacterial activity reflected the initial rate of leakage of K+ ions, suggesting that damage to cell membranes might be one of the major modes of action of these terpene alcohols [22]. The results also demonstrated that the initial rate of leakage and the amount of leaked K+ ions are useful as indices of the antibacterial activities of hydrophobic compounds [23]. The antimicrobial mechanism of totarol was studied using P. aeruginosa IFO 3080. This diterpene inhibited oxygen consumption and respiratory-driven proton translocation in whole cells, and oxidation of NADH in membrane preparation. NADH-cytochrome c reductase was inhibited by totarol while cytochrome c oxidase was not [22]. NADH-DPIP reductase and NADHCoQ reductase were also inhibited. The site of respiratory inhibition of totarol was thought to be near CoQ in the bacterial electron transport chain [21].
All the hydrolysable tannins tested demonstrated dose dependent membrane-damaging activity [22]. However, it remains to be elucidated whether their membrane-damaging activity directly contributes to their antibacterial action [22]. Carvacrol is a component of several essential oils and has been shown to exert antimicrobial activity [22]. It has been proved that carvacrol interacts with the membranes of bacteria such as B. cereus by changing its permeability for cations like H [+] and K [+] [22]. The dissipation of ion gradients leads to impairment of essential processes in the cell and finally to cell death [22]. Farnesol increased beta-lactam susceptibility of methicillin susceptible S. aureus by inhibition of cell wall biosynthesis through reduction of free C55 lipid carrier with subsequent retardation of murein monomer precursor transport across cell membrane [22].
Discussion and Conclusion
The use of herbal medicines in Asia represents a long history of human interactions with the environment [22]. Plants used for traditional medicine contain a wide range of substances that can be used to treat chronic as well as infectious diseases [21]. Due to the development of adverse effects and microbial resistance to the chemically synthesized drugs, people turned to ethnopharmacognosy [22]. They found literally thousands of phytochemicals from plants as safe and broadly effective alternatives with less adverse effect [22]. Many beneficial biological activities such as anticancer, antimicrobial, antioxidant, antidiarrheal, analgesic and wound healing activity were reported. In many cases the people claim the good benefit of certain natural or herbal products [22].
Pharmacognosy research confirms that plants can provides an effective bioactive product through submitting plant extracts to chemical investigation like LC and MS [23]. These techniques provide the structural information which in turn help to discover new bioactive compound present in new species of plants [23]. Likewise, by using new techniques and methods, we should find solutions to treat the disease caused by these deadly MDR pathogens and keep us ready for any new diseases or strains.
References
- Howard A, Michael D, Audrey F, Roy D (2012) Acinetobacter baumannii: An emerging opportunistic pathogen. Virulence 3(3): 243-250.
- Boundless (2016) Boundless microbiology, Boundless, America, pp. 775-4905.
- MRSA Tracking (2016) Center for disease control and prevention.
- Antibiotic resistance threats in the United States (2013) Center for disease control and prevention. Center for disease control and prevention.
- Handwerger, B Raucher, Altarac D, Monka J, Marchione S, et al. (1993) Nosocomial outbreak due to Enterococcus faecium highly resistant to vancomycin, penicillin, and gentamicin. Clinical Infectious Diseases 16(6): 750-755.
- Kau A, Steven M, William L, Ericka H, Michael G, et al. (2005) Enterococcus faecalistropism for the kidneys in the urinary tract of C57BL/6J Mice. American Society for Microbiology 73(4): 2461-2468.
- Chambers HF (2009) Waves of resistance: Staphylococcus in the antibiotic era. Nat Rev Microbiol 7(9):629-641.
- Allen NE, Nicas TI (2003) Mechanism of action of oritavancin and related glycopeptide antibiotics. FEMS Microbiol Rev 26(5): 511-532.
- Howard A, Michael O, Audrey F, Roy D, et al. (2012) Acinetobacter baumannii: An emerging opportunistic pathogen. Virulence 3(3): 243-250.
- Adzitey F (2015) Antibiotic classes and antibiotic susceptibility of bacterial isolates from selected poultry; a mini review. World Vet J 5(3): 36-41.
- Abraham E (1987) Cephalosporins. Drugs 4(2): 1-4.
- Bozdogan B, Appelbaum PC (2004) Oxazolidinones: Activity, mode of action, and mechanism of resistance. Int J Antimicrob Agents 23(2): 113-119.
- Epe B, Woolley P (1984) The binding of 6-demethylchlortetracycline to 70S, 50S and 30S ribosomal particles: A quantitative study by fluorescence anisotropy. EMBO J 3(1): 121-126.
- Gilbert D (2000) Aminoglycosides. In: Mandell GL, Bennett JE, Dolin R (Eds.), (5th edn), Mandell, Douglas, and Bennett's Principles and Practice of Infectious Diseases. Churchill Livingstone, London, UK, pp. 307-336.
- Wright GD (2010) Q and A: Antibiotic resistance: Where does it come from and what can we do about it? BMC Biol 8: 123.
- Holten KB, Onusko EM (2000) Appropriate prescribing of oral beta-lactam antibiotics. Am Fam Physician 62(3): 611-620.
- Beauregard DA, Williams DH, Gwynn MN, Knowles DJ (1995) Dimerization and membrane anchors in extracellular targeting of vancomycin group antibiotics. Antimicrob Agents Chemother 39(3): 781-785.
- Denyer SP, Hodges NA, German SP (2004) Introduction to pharmaceutical microbiology. Pharmaceutical Microbiology. In: (7th edn), Blackwell Science, UK, pp. 3-8.
- Gualerzi CO, Letizia B, Enrico C, Anna L, Roberto S, et al. (2000) Translation initiation in bacteria. The Ribosome: Structure, Function, Antibiotics, and Cellular Interactions. ASM Press, Washington, USA, pp. 477-494.
- Yixi X, Weijie Y, Fen T, Xiaoqing C, Licheng Ren (2015) Antibacterial activities of flavonoids; structure-activity relationship and mechanism. Current Medicinal Chemistry 22(1): 132-149.
- Saleem D, Pardi V, Murata RM (2017) Review of flavonoids: A diverse group of natural compounds with anti-candida albicans activity in vitro. Archives of Oral Biology 76: 76-83.
- Wang TY, Li Q, Bi KS (2017) Bioactive flavonoids in medicinal plants: Structure, activity and biological fate. Asian Journal of Pharmaceutical Sciences 13(1): 12-23.
- Thomas T (2013) Isolation, purification, and characterization of antibacterial principle from Drynaria Quercifolia.
- Leach KL, Steven M, Jerry R, William G, James R, et al. (2007) The site of action of oxazolidinone antibiotics in living bacteria and in human mitochondria. Mol Cell 26(3): 393-402.
- White D, Cox E (2013) Fighting the impact of antibiotic-resistance. FDA Consumer health Information.
- Henry RJ (1943) The mode of action of sulphonamides. Bacterial Rev 7(4): 175-262.
- Douthwaite S (1992) Interaction of the antibiotic’s clindamycin and lincomycin with Escherichia coli 23S ribosomal RNA. Nucleic Acids Res 20(18): 4717-4720.
- Hong W, Zeng J, Xie J (2014) Antibiotic drugs targeting bacterial RNAs. Acta Pharm Sin B 4(4): 258-265.
© 2022 Umesha S. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






