- Submissions

Full Text
Cohesive Journal of Microbiology & Infectious Disease
Highly Infectious, Less Pathogenic and Antibody Resistant Omicron Xbb.1, Xbb.1.5 and Xbb.1.5.1-Xbb.1.5.39 Subvariant Coronaviruses Do Not Produce Orf8 Protein Due To 8th Codon Gga=Tga Termination Codon Mutation
Asit Kumar Chakraborty*
Department of Biotechnology and Biochemistry, Vidyasagar University, India
*Corresponding author: Asit Kumar Chakraborty, Department of Biotechnology and Biochemistry, Oriental Institute of Science and Technology-West Bengal, Vidyasagar University, India
Submission: June 13, 2023; Published: August 03, 2023

ISSN 2578-0190 Volume6 issues5
Abstract
RNA viruses are very mutation prone. We recently reported SARS-CoV-2 ORF8 gene CAA=TAA and AAA=TAA Termination Codon Mutations in B.1.1.7 variants with no production of viable ORF8 protein. We described here another GGA=TGA termination codon in the 8th codon of ORF8 gene located exclusively in XBB.1 (XBB.1.16 and XBB.1.22) and XBB.1.5 subvariants (XBB.1.5.1 to XBB.1.5.39) but not in XBB.2 variant or Alpha, Beta, Gamma, Delta and Omicron BA.1, BA.2, BA.4, BA.5, BF.7 and BQ.1 subvariants. However, G>T mutation at 27915 also created an alternate ATG codon but the protein product was short due to preceding TAG and TGA termination codons. The originally located following ATG codons were there but in alternate reading frames and ultimately no ORF8 protein was formed in XBB.1.5 subvariants which were spreading highly now over BA.2.75, BA.4.6, BA.5.2.1, BF.7 and BQ.1.1 subvariants. This is a vivid example of three termination codon mutations in the coronavirus ORF8 protein which was implicated as target for many human proteins regulating interferon production, chromosome instability, antibody production, patho genicity and virus clearance. The 30nt deletion in the 3’-UTR, including 24LPP, and 145Y deletions in Spike protein as well as N-protein 31ERS and ORF1ab polyprotein 3675SGF deletions made XBB.1.5 Omicron coronavirus weak and less pathogenic so that WHO declared coronaviruses as non-emergency pathogen.
Keywords:Termination codon mutants; ORF8 protein; COVID-19; Immunomodulation; Higher transmission; Lower pathogenicity
Introduction
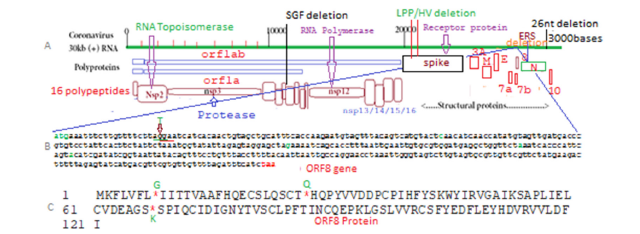
The SARS-CoV-2 is the causative agent of the coronavirus disease 2019 with severe public health consequences and million deaths [1-9]. The novel SARS-CoV-2 shares nearly 96% similarity to the bat coronavirus isolate RaTG13, suggesting these animals are the likely natural reservoir of the virus [10,11]. Thus, different animal, birds and whale corona viruses were known since 2003 but human corona viruses were appeared in December 2019 at Wuhan province of China. So far million coronaviruses RNA were sequenced and divided into Alpha, Beta, Delta, Gamma etc. as well as Omicron variants with mutations, deletions and insertions [12-18]. There were many differences in pathogenic potential and immunogenicity among those VOCs. SARS-CoV-2 is a large positive-stranded RNA virus with round 30000 nucleotides genome (Figure1). It has structural proteins Membrane (M), Envelope (E), Nucleocapsid (N), Spike (S) coded from 3’-1/3 part of the virus independently, but RNA-dependent RNA polymerase was coded from nsp12 domain of ORF1ab polyprotein coded from 2/3 of the 5’-parts of the genome and such polyprotein was degraded into sixteen polypeptides (nsp1- nsp16). The nsp2 protein is RNA topoisomerase with new therapeutic target whereas Nsp3 and nsp5 are proteases that cleave polyprotein into sixteen polypeptides with diverse functions [18-23]. The nsp6, nsp7, nsp8, nsp9 and nsp10 were small accessory proteins involved in RNA polymerase replication complex. The nsp14 and nsp15 are ribonucleases and nsp16 is 2’-O methyltransferase and nsp13 is RNA helicase and may have capping methyl transferase activity. The ORF3a, ORF6, ORF7a/7b, ORF8 and ORF10 small proteins also coded from 3’ end of the genome and have roles in regulating cellular genes.
Figure 1:Structure of Omicron corona virus with polypeptides (A), nucleotide sequence of ORF8 gene (B) and ORF8 protein sequence with three termination codons detected (C).
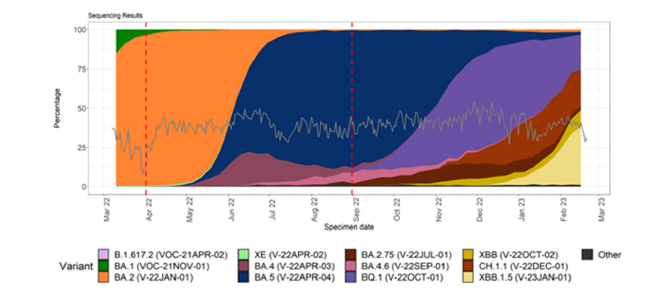
In USA, Wuhan-D614G mutant first peak between March- August 2020, Alpha (B.1.1.7) 2nd peak with spike 69HV deletion immune-escape mutant between January-June 2021 followed by 3rd peak of Delta (B.1.617.2, AY.X) with spike 157FR deletion mutant between June to December 2021 [24]. Since last week of December 2022 4th peak of Omicron BA.1 variant (B.1.1.519) spread was evident followed by BA.2 variant spread since April 2022 with 29 mutations in the spike. From June-July 2022, Omicron BA.4/BA.5 variants were dominating worldwide followed BF.7, BQ.1, BQ.1.1, XBB.1 and XBB.1.5 subvariants recently (Figure 2).
Figure 2:Distribution and sequencing data comparison for COVID-19 in the United Kingdom.
The Spike protein (1273AA) of COVID-19 had gone extensive mutations and deletions than large polyprotein ORF1ab (7096aa) particularly in Omicron lineages. The spike 24LPP, 69HV, 143VYY, 157FR, 212L and 145Y deletions were detected in different proportion whereas 215EPE and 249RWMD insertions were also reported in Omicron BA.1 variant and BQ.1 variant respectively [25-29]. Among the ORF1ab deletions, 141KSF deletion in nsp1 domain was found only in omicron BA.4 subvariants and 3674LSG deletion found in omicron BA.1 subvariant whereas 3675SGF deletion in nsp6 domain was found in most Omicron (BA.1, BA.2, BA.4, BA.5) and Alpha (B.1.1.7) variants. Dominant point mutations D614G and N501Y were important for higher transmission whereas ~20 mutations in the RBD domain of Omicron were not found in deadly B.1.1.7, B.1.617.2 and AY.103 variants. The E484A, T478K, L452R and K417N/T mutations were very immune-modular and such mutant viruses were refractory to antibodies of patients.
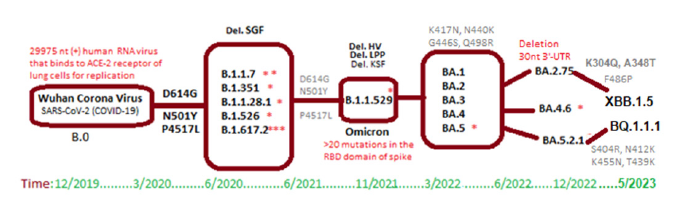
More than a few dozen spike mutations were recently detected in XBB.1.5 and BQ.1.1.1 subvariants. However, Omicron variants were less pathogenic and usually did not require oxygen support and hospitalization unless co-morbidity [30]. Still pneumonia, chest pain, confusion, and headache with cough and cold, were different symptoms that affected over 650 million people worldwide. The gradual changes in different SARS-CoV-2 variants since 2019 was shown in Figure 3. Presently, infectivity of XBB.1.5 subvariant was dominating worldwide but very mild symptoms due to huge mutations in spike and deletion of 26nt 3’-UTR. However, the titer of such deleted coronaviruses was low and their spread may be due to better interaction with ACE-2 receptor and reinfections of patients [31-36].
Figure 3:Conversion of Wuhan B.0 coronavirus into Omicron viruses like BQ.1.1.1 and XBB.1.5.3 types of sub subvariants.
Accumulating evidence suggested that small regulatory proteins (ORF3a, ORF7a, ORF8) of SARS-CoV-2 interacted highly with many cellular proteins. Preliminary 3-D graphics interactive studies indicated few dozen proteins like PVR, IRF3, ATF6, Beclin 1, FK506-binding protein 10, EDEM, vitronectin, OPJ94, Sec62163, VIP36, TRFT3 and PLAT etc interacted with ORF8 protein regulating protein folding, apoptosis and interferon production. Such process likely favours COVID-19 survival in host cells inhibiting immune control mechanisms [37]. Genetic analysis pointed a severe deletion in ORF8 (Δ382) caused less severe corona infections likely due to low viral load with increased immune clearance.
However, in cell culture study with such deletion mutant contradicted the finding of lower viral load with no change of cellular transcriptional profile. The ORF8 protein also mediates immune evasion by downregulating MHC-I molecules like HLA-A2 [38]. The IgG domains similarity of ORF8 protein may be important to modulate host immune functions and chromatin structure. The C>T base change at 27972nt and another A>T base change at 28095nt created two termination codons (CAA=TAA and AAA=TAA) to produce 26AA and 67AA long ORF8 truncated proteins. Similar Blast-N search with mutated oligonucleotides detected many ORF8 mutants with distinct S24L, V32L, P38S, R52I, A65V, Y73C, L84S, K92E and V100L mutations with or without TAA termination mutations [39].
During our database search to characterize the XBB.1.5 lineages, we noticed no expression data for ORF8 protein in the genomic sequences. As we already known the two termination codons in ORF8 gene of Alpha variants, we searched the similar termination codon mutation in XBB-related variants which originated from BA.2.10 and BA.2.75 recombination and mutation [39-46]. We detected here a new termination codon mutation in ORF8 gene of XBB.1 lineage but not in XBB.2 lineage. Spike protein mutations and deletions had affected Omicron coronaviruses and for transformation of BA.2 to BA.2.75 required K147E, W152R, F157L, I210V, G257S, D339H, G446S, N460K and Q493R mutations in the spike protein and BA.2.75 was originated in 31.12.2021. Gene rearrangement and deletion in the 5’-UTR and 3’-UTR were also demonstrated in different SARS-CoV-2 variants. The article was deposited in Research Square preprint on 30th May 2023.
Methods
We searched PubMed to get an idea on published papers on ORF8 and searched SARS-CoV-2 NCBI database using BLAST-N and BLAST-X search methods. Multi-alignment of protein was done by Mult Alin software and multi-alignment of DNA by CLUSTAL-Omega software. 1st impression of ORF8 mutants was gained by Blast N searching of deletion boundary of 120nt sequence and analyzing the sequences with 90-100% similarities. Blast X search of ORF8 full length gene used to get mutant ORF8 proteins with or without termination codon. Then, the other ORF8 mutants were detected by Blast-N search of TAA mutant oligos as well as other oligos selected from point mutation boundaries [47,48]. The hairpin structure of ORF8 gene 222nt 5’-terminal sequence was done by Oligo Analyzer 3.1 software (Integrated DNA Technologies). The protein 3-D structure was determined by SWISS-Model software [49-61].
Result
In Table 1, we showed XBB.1.5. subvariants specific changes in the coronavirus proteins whereas K304Q and A411S two important mutations in the RBD of spike of XBB.1.5.3 sub-subvariant might be significant. Similarly, XBB.1.5.29 and XBB.1.5.30 had A348V and A348T mutations in the spike but roles of such mutations had to be tested yet. The never-the-less dominant common F486P spike mutation was implicated in antibody evasion and higher transmission in XBB.1.5 subvariants. We found six mutations (P2045S, T2137A, A3697V, T5941I, H5951Y, P6376S) in ORF1ab polyprotein of XBB.1.5.30 subvariant and four different mutations (S1188L, P2045L, P2110S, N6481K) in XBB.1.5.20 which also harboured A398V mutation in the N-protein (Table 1). Three spike mutations (P463S, E554K, P1162S) in XBB.1.5.8 might be significant and the penetration of such subvariant in the database was low. Similarly, four spike mutations (V83S, Y200C, V382L, T573I) in the XBB.1.22.1 subvariant was reported in Table 1. Interestingly, we detected total eight mutations in N-protein of XBB.1.5.sub-subvariants: R10Q in XBB.1.5.8, G25C in XBB.1.5.16, D81H in XBB.1.5.38, I131R in XBB.1.5.3, R195I in XBB.1.5.19, P279L in XBB.1.5.28, S327L in XBB.1.5.9 and A398V in XBB.1.5.20. Mutation rate was higher in recent coronavirus isolates (BQ.1.1.1 and XBB.1.5.1) and we also found four distinct mutations in ORF3A trans-activator protein: G49C in XBB.1.5.21, S92L in XBB.1.5.2, G172D in XBB.1.5.38 and H182Y in XBB.1.5.27 [61-68].
Table 1:Demonstration of major mutations in Omicron XBB.1.5.1-XBB.1.5.39 sub subvariants. XBB.1.5.26 (OQ783732) and XBB.1.5.31 (OQ758970) have no mutation in our search as stated here (ORF1ab, Spike, N, M, E, Orf3A) and also other small ORF proteins. No M protein was conserved but A63T mutation was found in all omicron variants and a D3G mutation in BA.1 variant and a I82T mutation in Iota variant were detected.

After the demarcation of XBB.1.5 sub-subvariants, we did multi-alignment analysis to pinpoint the genetic changes and if ORF8 termination codon mutation happened in all those subsubvariants. For multi-alignment, we choose B.o, B.1.1.7, B.1.617.2, BF.7, BA.2.75 and BQ.1 as standard variant and subvariant whereas few XBB.1.5 sub-subvariants as experimental [69]. In truth, we checked all XBB.1.5.1 to XBB.1.5.39 sub-subvariants for negative ORF8 expression in the database. Figure 4 stated that ORF8 gene mutation (GGA=TGA) in all XBB.1.5.1 sub-subvariants but not in XBB.2 as well as standard variants. We put standard CAA=TAA termination codon mutant (accession no. OP711844) and standard AAA=TAA termination codon mutant (accession no. OP683545) belonging to Alpha (B.1.1.7) variant for comparison and all three termination codon mutants did not produce viable ORF8 protein to interact and modulate host genes involved in interleukins expression and immune modulation [70-75].
Figure 4:Localization of three termination codon mutants in ORF8 gene of SARS-CoV-2.

We checked the 3675SGF dominant deletion in the nsp6 domain of ORF1ab polyprotein and except in Wuhan and Delta, all Omicron (BA.1/2/4/5, BF.7, BQ.1, XBB.1) and Alpha (B.1.1.7) lineages had such deletion (Figure 5). Similarly, we checked the nsp1 deletions in XBB.1.5 and no such deletion was found (Figure 6). Astonishingly, we detected a B.1.1.7 alpha variant with GHVMV deletion which also had ORF8 termination codon mutation (accession no. OP711844). Next, we determined if there was any Spike deletion in XBB.1.5 subvariants. Spike 24LPP deletion was found in BA.2 lineages including BA.2.75, XBB, XBB.1, XBB.1.5 (Figure 7) but no 69HV deletion. On the contrary, both 24LPP and 69HV spike deletions were found in BQ.1, BF.7, BA.4/5 but not in Delta variant whereas only 69HV deletion found in Alpha variant. We also demonstrated that BA.2.75, XBB, XBB.1 and XBB.1.5 subvariants had N501Y mutation (Figure 8) and D614G mutation both of which increased transmission to over 100% than Wuhan virus.
Figure 5:Demonstration of all omicrons including XBB, XBB.1, XBB.2 and XBB.1.5 subvariants as well as Alpha variant had SGF deletion in nsp6 protein but Delta and Wuhan.

Figure 6:Demonstration of absence of GHVMV and KSF deletions in nsp1 protein of XBB.1.5 subvariants.

Figure 7:Demonstration of Spike deletion in XBB.1.5 subvariants. Spike 24LPP deletion found in BA.2 lineages including BA.2.75, XBB, XBB.1, XBB.1.5 (A) but no spike 69HV deletion (B). The 69HV deletion was also found in Alpha, BQ.1, BF.7, BA.4 but not in Delta variant.
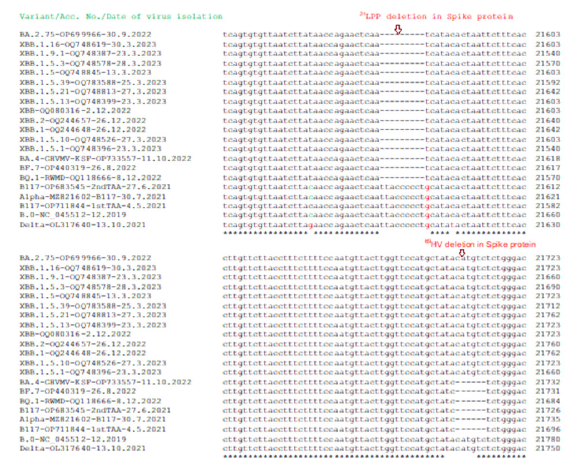
Figure 8:Multi-alignment demonstration that genomes of BA.2.75, XBB, XBB.1 and XBB.1.5 subvariants had N501Y mutation similar to B.1.1.7 (A) and D614G mutation similar to B.1.1.7, B.1.617.2 and others (B). Delta variant had D614G mutation but not N501Y mutation. Wuhan related early corona viruses (B.0, B.1, B.1.1) had no both D614G and N501Y mutations.

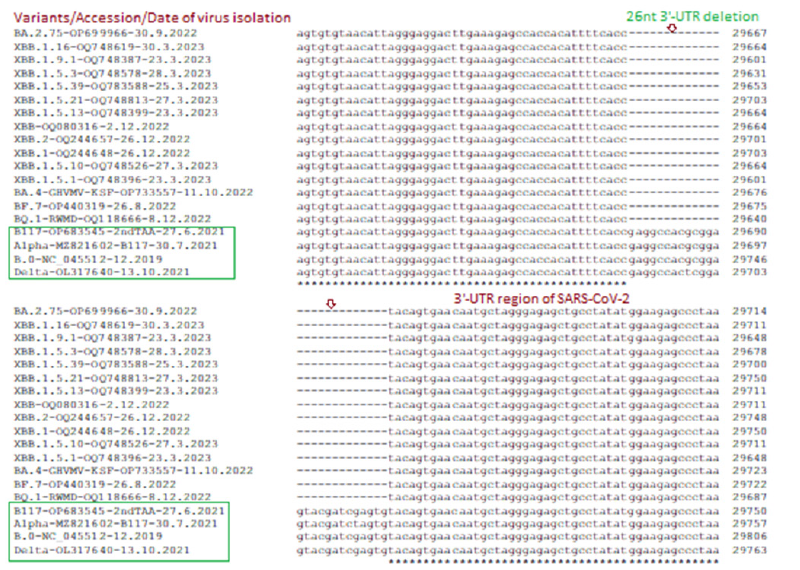
The deadly Delta varianN501YG mutation but no N501Y mutation whereas control 2019 Wuhan virus (B.0) had no D614G and N501Y both mutations. Previously, we showed that N-protein 31ERS three amino acids deletion was prominent in Omicron variants starting from B.1.1.529 variants including BA.2, BA.2.9, BA.2.75 and BA.2.75.2 [76-78]. However, Wuhan, BA.1.1, B.1.1.1, BA.1.1.172, B.1.1.7, B.1.617.2 and other early coronavirus lineages had no such deletion. Figure 9 showed that BA.4, BA.5, BF.7, BQ.1, XBB.1 lineages also had such deletions which caused N-protein three amino acids shorter (216AAs). Most importantly, previously we reported 26nt deletion in 3’-UTR in many Omicron lineages (BA.2, BA.4, BA.5) which made coronavirus very replication defective. Interestingly, we did not find 26nt deletion in BA.1 early omicron lineages or old Wuhan, Alpha, Beta, Gamma, Delta coronavirus variants. In Figure 10, we showed that XBB.1.1 lineages retained such deletion including XBB.1, XBB.2, XBB.1.16, XBB.1.22.1, BF.7, BQ.1, BQ.1.1 and BQ.1.1.1 whereas spread of such variant likely increased than BA.2.75, BF.7 and BQ.1.1.1 subvariants.
Figure 9:Multi-alignment demonstration of 31ERS N-protein deletion in Omicron corona viruses but not in Alpha, Delta or Wuhan early coronaviruses. The other early VOCs like Beta, Gamma, Epsilon, Zeta also had no such deletion (data not shown).
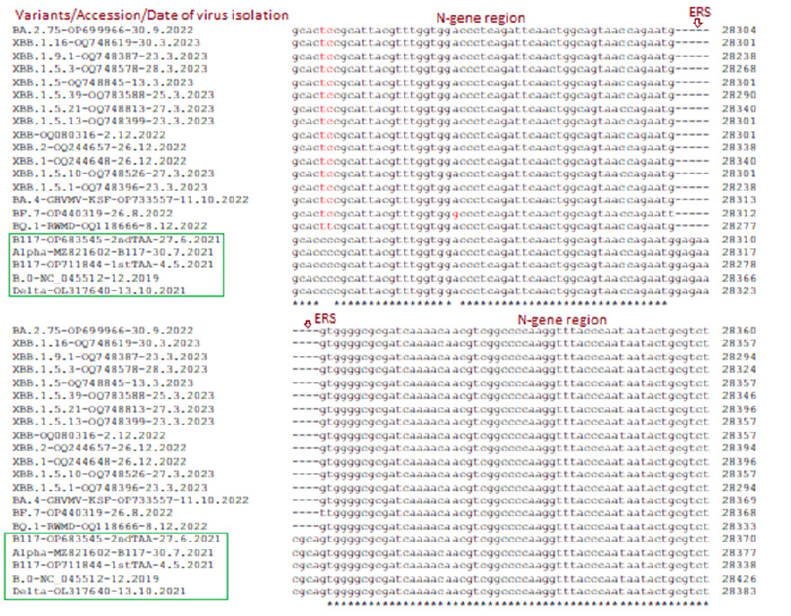
Figure 10:Multi-alignment demonstration of 26nt 3’-UTR deletion in most Omicron coronaviruses (BA.2.75, BA.4, BF.7, BQ.1, XBB.1.5.1) but such deletion was not reported in Alpha, Delta and Wuhan early coronaviruses.
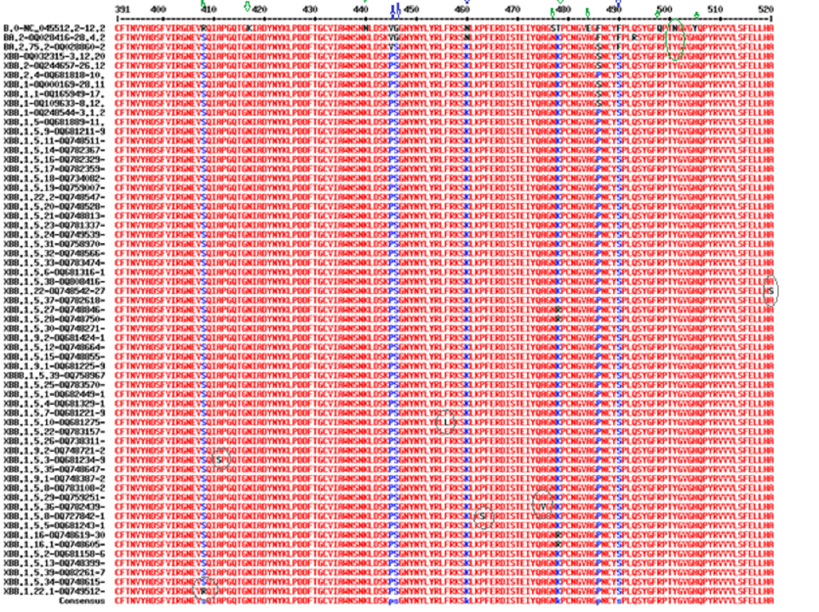
We showed the XBB.1.5.1 to XBB.1.5.39 specific mutations in most COVID-19 proteins in Table 1. We showed the part of the multi-alignment of spike protein RBD domain in Figure 11. Figure 11 also demonstrated Omicron BA.2 specific mutations (green arrows), XBB.1.5 specific mutations (blue arrows), N501Y mutation (green circle) and XBB.1.5.1 sub subvariants mutations (grey circles). Further, A411S (XBB.1.3), P463S (XBB.1.5.8), F456l (XBB.1.5.10), A475V (XBB.1.5.36) including A520S (XBB.1.22) and S408W (XBB.1.22.1) sub subvariant specific mutations were demonstrated (Table 1). Those sub sub variant specific mutations will be utilized to make subvariant specific oligonucleotides for the detection of unknown COVID-19 variants as well as to demonstrate the database penetration of those sub subvariants by BLAST search.
Figure 11:Multi-alignment of spike protein RBD domain region to demonstrate Omicron BA.2 specific mutations (green arrows), XBB.1.5 specific mutations (blue arrows), N501Y mutation (green circle) and XBB.1.5.1 sub subvariants mutations (grey circles).
Discussion
Five VOCs of SARS-CoV-2 mainly caused million deaths worldwide and named as B.1.1.7 (U.K.), B.1.351 (South Africa), P.1 (Brazil), B.1.617.2 (India), and B.1.1.529 (Africa). The B.1.1.529 variant was named Omicron which diverged into different BA.1, BA.2, BA.3, BA.4 and BA.5 lineages worldwide during 2022 with mild infections. Viral transactivator proteins regulate cellular genes to modulate immunogenicity and pathogenicity. As for example, in HIV retrovirus mediated pathogenesis, TAT, NEF and REV small proteins modulate its own transcription as well as human cellular genes. Similarly, preliminary reports indicated that corona virus ORF8 protein (121 AAs) acts as histone mimics disrupting chromatic structure with many epigenetic changes and immune modulator functions. ORF8 protein could inhibit MHC-1 and IFNbeta functions due to some similarities to immunoglobulin domains and also modulate spike protein. A 382-nucleotide deletion (Δ382) in the ORF8 region of the corona virus genome causes weak virus load and weak pathogenicity (accession no.MT374101) [78-83]. Previously, we showed that C>T base change at 27972nt and another A>T base change at 28095nt created two termination codons (CAA=TAA and AAA=TAA) to produce 26AA and 67AA long ORF8 truncated proteins. Further, S24L, V32L, P38S, R52I, A65V, Y73C, L84S, K92E and V100L mutations in the ORF8 gene located with or without TAA termination mutations. Thus, ORF8 gene is very proven to mutation disrupting its function which controls immunogenicity and virus clearance.
Over time, the coronavirus has undergone mutations and deletions and different variants reported in different parts of the world with different time since December 2019. We found no GGA=TGA termination codon mutation in Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1/B.1.1.28.1), Delta (B.1.617.2), Kappa (B.1.617.1), Epsilon (B.1.427/B.1.429), Zeta (P.2); Eta (B.1.525), Iota (B.1.526) and B.1.1.298 (Mink Variant). Spike mutations are of major health concerns, as they reportedly exacerbate the infectious rate of the virus as in D614G and N50Y mutations. We found here major ORF8 truncated mutant in XBB.1 and XBB.1.5 lineages which were rapidly spreading now. Interestingly, RNA recombination generated few omicron lineages similar to XBB which was generated due to recombination between BA.1.10 and BA.2.75. Thus, XBB to XBB.1 and then XBB.1 to XBB.1.5 may be important but we characterized up to XBB.1.5.39 and in all we detected GGA=TGA termination codon mutation.
Thus, present outbreaks occurred with a corona virus with ORF8 gene product deficient. We also know that in most Omicron corona viruses, a 26nt deletion was prominent and we do not find any report that an insertion in that 3’ UTR region has happened yet in Omicron lineages! Dominant mutation and deletion of small regulatory protein ORF7a limits viral suppression of the interferon response. We recently reported such deletions abolishing the production of both ORF7a and ORF7b proteins (see, accession no. OP711842). A WA1/BA.5 bivalent mRNA vaccine was inactive to kill BQ.1 and XBB.1 coronaviruses and titre against BQ and XBB subvariants were lower by 13-81fold and 66-155fold respectively as compared to Omicron BA.2 and BA.5 variants. Antiviral COVID-19 medications such as Paxlovid (Nirmatrelvir/Ritonsvir) and Remdesivir should still be effective against XBB.1.5 variant and both of these drugs will prevent virus replication as RdRp enzyme crucial function is not altered in XBB.1.5 [29,37].
The most sequences in SARS-CoV-2 database we analyzed were from USA (Howard, D. et al group) and we characterized some Bangladesh omicron data which confirmed that GGA=TGA termination codon was found in XBB.1 variant (accession no. OQ075411) but not in XBB variant (accession no. OQ075381) or XBB.2 variant (accession no. OQ075400). Interestingly, we also recently pinpointed round 400 147RWMD spike insertion mutants in US patients. We found three Northern Ireland (UK) based patients where such RWMD insertion was observed (accession numbers; OX527225, OX520545, OX486753). The 5’-UTR of SARS-CoV-2 forms five hairpin structures and nsp1 protein bound to first stem and loop structure helping transport of nsp1 to ribosome. Thus, 26nt 3’-UTR (29534-29870nt) S2M long stem-loop structure implicated in replication and other function in the coronavirus biology. The size of the genome has been changed from 29903nt to 29782nt. Thus, point mutation, deletion and insertion greatly affected coronavirus biology and WHO declared that recent isolates were no more so dangerous to cause death in COVID-19 infected individual [31,83].
HIV transactivator protein, TAT (71 AAs) binds to promoters of genes that are bound by the ETS1 transcription factor, the CBP histone acetyltransferase suggesting its role in regulating genes involved in T cell biology and immune response. Tat protein binds to TAR region where other cellular protein (puralpha) also docks. Further, Tat binds to breast cancer resistant protein implying its role in cancer pathogenesis. Nef protein (216 AAs) activates cellular Src and Tec tyrosine kinases after binding Nef-dimer to SH3-SH2 domain of tyrosine kinases activating transcription of HIV. The Rev protein (129 AAs) was suggested as HIV RNA transporter and such function was inhibited by nuclear factor 90, a dsRNA binding host protein whereas other host proteins XPO1 (CRM1) and RBM14 may be involved [9,69].
Strikingly, ORF8 protein 62-77 residues has Ig-like domain and binds CD16a (FcγRIIIA) with high affinity. Such interaction lowers the activity of CD16 at the surface of monocytes and NK cells reducing the capacity of PBMCs and monocytes to mediate antibody-dependent cellular cytotoxicity and humoral responses The crystal structure of SARS-CoV-2 ORF8 reveals a ∼60-residue core polypeptides has potential similarity to ORF7a interface containing two dimerization interfaces and a covalent disulfidelinked dimer through an N-terminal sequence, while a separate noncovalent interface is formed by a carboxy-terminal SARS-CoV- 2-specific sequence, 73YIDI76 . We found many deletions in the ORF7a protein also and thus both should be transactivator proteins absence of which markedly lower the pathogenicity. Unfortunately, although ORF7a and ORF8 NH2-terminal 40 AAs has 25% scattered similarities, no such minor similarity with TAT protein or other HIV transactivator proteins like Nef and Rev (data not shown). Never-the-less, the roles of ORF8 protein in COVID-19 replication and pathogenesis were emerging and absence of ORF8 protein due to termination codon mutation was now well established in XBB.1.5 and B.1.1.7 variants. Thus, translational suppression of ORF8 protein is a therapeutic method to control Corona virus pathogenicity [29,37].
Conclusion
Presently circulating XBB.1.5 sub subvariants (XBB.1.5.1- XBB.1.5.39, XBB.1.16, XBB.1.22, XBB.1.9.1, XBB.1.9.2 etc) have GGA=TGA termination codon mutation in the ORF8 gene and such ORF8-deficent coronaviruses are less pathogenic. The 26nt deletion in the 3’-UTR, 24LPP and 140Y deletions in spike and 31ERS deletion in N-protein may be significant in such a process.
Acknowledgement
We thank NCBI (NIH, USA) for the free SARS-CoV-2 database. Free CLUSTAL-Omega software is also acknowledged.
References
- Akaishi T (2022) Insertion and Deletion mutations between the genomes of SARS-CoV, SARS-CoV-2, and bat coronavirus RaTG13. Microbiol Spectr 10(3): e00716-22.
- Ali A, Mishra R, Kaur H, Chandra BA (2021) HIV-1 Tat: An update on transcriptional and non-transcriptional functions. Biochimie 190: 24-35.
- Alkhansa A, Lakkis G, Zein LE (2021) Mutational analysis of SARS-CoV-2 ORF8 during six months of COVID-19 pandemic. Gene Rep 23: 101024.
- Altschul SF, Gish W, Miller W, Myers EM, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol. 215: 403-410.
- Aryal M, Lin D, Regan K, Shoucheng D, Haibin S, et al. (2022) The HIV-1 protein Nef activates the tec family kinase Btk by stabilizing an intermolecular SH3-SH2 domain interaction. Sci Signal 15(752): eabn8359.
- Badua CL, Baldo K, Medina P (2021) Genomic and proteomic mutation landscapes of SARS‐CoV‐ J Med Virol 93(3): 1702-1721.
- Beaudoin B, Arduini A, Bourassa C, Medjahed H, Chen L, et al. (2022) SARS-CoV-2 accessory protein ORF8 decreases antibody-dependent cellular cytotoxicity. Viruses 14(6): 1237.
- Biswal M, Diggs S, Xu D, Nelli K, Jiuwei L, et al. (2021) Two conserved oligomer interfaces of NSP7 and NSP8 underpin the dynamic assembly of SARS-CoV-2 RdRP. Nucleic Acids Res 49(10): 5956-5966.
- Budhiraja S, Liu H, Couturier J, Malovannaya A, Qin J, et al. (2015) Mining the human complexome database identifies RBM14 as an XPO1-associated protein involved in HIV-1 rev function. J Virol 89(7): 3557-3567.
- Chakraborty AK (2023a) The 249RWMD spike protein insertion in Omicron BQ.1 subvariant compensates the 24LPP and 69HV deletions and may cause severe disease than BF.7 and XBB.1 subvariants. Research Square.
- Chakraborty AK (2023b) The 82GHVMV and 141KSF deletions in the Nsp1 protein of ORF1ab polyprotein favour the creation of immune-weak SARS-CoV-2. Sun Text Rev Virol 3(2): 137.
- Chakraborty AK (2023c) Conversion of B.0 lineage of human corona virus (Covid-19) into notoriously infecting less Pathogenic and Immune Escape Omicron B.1.1.529.2.75.2 or BA.2.75.2 Variant. Journal of BioMed Research and Reports 2(1).
- Chakraborty AK (2022a) Hyper-variable spike protein of omicron corona virus and its differences with alpha and delta variants: Prospects of RT-PCR and new vaccine. J Emerg Dis Virol 7(1): 1-13.
- Chakraborty AK (2022b) A method of identification of SARS-CoV-2 variant using NCBI BLAST-2 100% homology search with specific oligonucleotides selected at the deletion boundaries of S, N, ORF7a, ORF8 and ORF1ab proteins. Research Square.
- Chakraborty AK (2022c) Dynamics of SARS-CoV-2 ORF7a gene deletions and fate of downstream ORF7b and ORF8 genes expression. SunText Rev Biotechnol 3(1): 142.
- Chakraborty AK (2022d) SARS-CoV-2 ORF8 gene CAA=TAA and AAA=TAA termination codon mutations found mostly in B.1.1.7 variant was independent of popular L84S mutations. Int J Clini Med Edu Res 1(6): 192-208.
- Chakraborty AK, Chanda A (2021) New biotechnological exploration on COVID-19 proteins: Functions, mutational profiles and molecular targets for drug design. Sun Text Rev Virol 2(1): 115.
- Chakraborty AK (2020a) Coronavirus Nsp2 protein homologies to the bacterial DNA Topoisomerase I and IV Suggest Nsp2 Protein is a unique RNA Topoisomerase with novel target for drug and vaccine development. Virol Mycol 9: 185.
- Chakraborty AK (2020b) Clinical, diagnostic and therapeutic implications of coronavirus ORFab polyprotein associated Nsp16 Protein-A bioinformatics approach. Acta Scientific Medical Sciences 4(5): 97-103.
- Chakraborty AK (2020c) Multi-alignment comparison of coronavirus non-structural proteins Nsp13-16 with ribosomal proteins and other DNA/RNA modifying enzymes suggested their roles in the regulation of host protein synthesis. International J Clini Med Informatics 3: 7-19.
- Chaudhari AM, Singh I, Joshi M, Patel A, Joshi C (2022) Defective ORF8 dimerization in SARS-CoV-2 delta variant leads to a better adaptive immune response due to abrogation of ORF8-MHC1 interaction. Mol Divers 27(1): 45-57.
- Chen J, Lu Z, Yang X, Zhou Y, Gao J, et al., (2022) Severe acute respiratory syndrome coronavirus 2 ORF8 protein inhibits type i interferon production by targeting HSP90B1 signalling. Front Cell Infect Microbiol 12: 899546.
- Chou JM, Tsai JL, Hung JN, Chen IH, Chen ST, et al. (2022) The ORF8 protein of SARS-CoV-2 modulates the Spike protein and Its implications in viral transmission. Front Microbiol 13: 883597.
- Chen X, Zhou Z, Huang C, Zhou Z, Kang S, et al., (2021) Crystal structures of bat and human coronavirus ORF8 protein ig-like domain provide insights into the diversity of immune responses. Front Immunol 12: 807134.
- Corpet F (1988) Multiple sequence alignment with hierarchical clustering. Nucl Acids Res 16(22): 10881-10890.
- Dandekar DH, Ganesh KN, Mitra D (2004) HIV-1 Tat directly binds to NFkappaB enhancer sequence: Role in viral and cellular gene expression. Nucleic Acids Res 32(4): 1270-1278.
- DeRonde S, Deuling H, Parker J, Chen H (2022) Identification of a novel SARS-CoV-2 variant with a truncated protein in ORF8 gene by next generation sequencing. Sci Rep 12: 4631.
- Dingwall C, Ernberg I, Gait MJ, Green SM, J Karn, et al. (1989) Human immunodeficiency virus 1 tat protein binds Trans-Activation-Responsive Region (TAR) RNA in vitro. Proc Natl Acad Sci 86(18): 6925-6929.
- Erukainure OL, Atolani O, Muhammad A, Ravichandran R, Musa M, et al. (2021) Translational suppression of SARS-COV-2 ORF8 protein mRNA as a Viable therapeutic target against COVID-19: Computational studies on potential roles of isolated compounds from Clerodendrum volubile leaves. Comput Biol Med 139: 104964.
- Fahmi M, Kitagawa H, Yasui G, Kubota Y, Masahiro IM (2021) The functional classification of ORF8 in SARS-CoV-2 replication, immune evasion, and viral pathogenesis inferred through phylogenetic profiling. Evol Bioinform 17: 11769343211003079.
- Farkas C, Mella A, Turgeon M (2021) A novel SARS-CoV-2 viral sequence bioinformatic pipeline has found genetic evidence that the viral 3′ Untranslated Region (UTR) is evolving and generating increased viral diversity. Front Microbiol 12: 665041.
- Flower TG, Buffalo CZ, Hooy RM, Allaire M, Ren X, et al. (2021) Structure of SARS-CoV-2 ORF8, a rapidly evolving coronavirus protein implicated in immune evasion. Proc Natl Acad Sci 118(2): e2021785118.
- Gamage AM, Tan KS, Chan WO, Liu J, Tan CW, et al. (2020) Infection of human nasal epithelial cells with SARS-CoV-2 and a 382-nt deletion isolate lacking ORF8 reveals similar viral kinetics and host transcriptional profiles. PLoS Pathog 16(12): e1009130.
- Gandhi L, Maisnam D, Rathore D, Preeti C, Anvesh B, et al. (2022) Respiratory illness virus infections with special emphasis on COVID-19. Eur J Med Res 27(1): 236.
- Gong YN, Tsao KC, Hsiao MJ, Chung G, Peng N, et al. (2020) SARS-CoV-2 genomic surveillance in taiwan revealed novel ORF8-deletion mutant and clade possibly associated with infections in Middle East. Emerg Microbes Infect 9(1): 1457-1466.
- Gordon DE, Hiatt J, Bouhaddou M, Rezeli VV, Ulferts S, et al. (2020) Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms. Science 370(6521): eabe9403.
- Goud VR, Chakraborty R, Chakraborty A, Lavudi K, (2022) A bioinformatic approach of targeting SARS-CoV-2 replication by silencing a conserved alternative reserve of the orf8 gene using host miRNAs. Comput Biol Med 145: 105436.
- Hassan SS, Choudhury PP, Dayhoff GW, Alaa AA, Vladimir N, et al. (2022a) The importance of accessory protein variants in the pathogenicity of SARS-CoV-2. Arch Biochem Biophys 717: 109124.
- Hassan SS, Kodakandla V, Redwan EM, Lundstrom K, Bruno S, et al. (2022b) An issue of concern: unique truncated ORF8 protein variants of SARS-CoV-2. Peer J 10: e13136.
- Hassan SK, Aljabali AA, Panda PK, Ghosh S, Adam M, et al. (2021) A unique view of SARS-CoV-2 through the lens of ORF8 protein. Comput Biol Med 133: 104380.
- Habib MT, Afrad MH, Rahman S, Khan MH, Firdausi Q, et al. (2023) Coding-complete genome sequences of 40 SARS-CoV-2 omicron XBB, XBB.1, and XBB.2 sublineage strains in Bangladesh. Microbiol Resour Announc 12(3): e0000123.
- Hachim A, Gu H, Kavian O, Mori M, Kwan MY, et al. (2022) SARS-CoV-2 accessory proteins reveal distinct serological signatures in children. Nat Commun. 13: 2951.
- Harvey WT, Carabelli AM, Jackson B, Gupta RK, David L, et al. (2021) SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol 19(7): 409-424.
- Kee J, Thudium S, Renner DM, Glastad K, Palozola K, et al. (2022) SARS-CoV-2 disrupts host epigenetic regulation via histone mimicry. Nature 610(7931): 381-388.
- Lasrado N, Collier AY, Miller J, Hachmann NP, Liu J, et al. (2023) Waning immunity against XBB.1.5 following bivalent mRNA boosters. Bio Rxiv 2023.
- Liu P, Wang X, Sun Y, Zhao H, Cheng F, et al. (2022) SARS-CoV-2 ORF8 reshapes the ER through forming mixed disulfides with ER oxidoreductases. Redox Biol 54: 102388.
- Lu G, Wang Q, Gao GF (2015) Bat-to-human: spike features determining ‘host jump’ of coronaviruses SARS-CoV, MERS-CoV, and beyond. Trends Microbiol 23(8): 468-478.
- Marban C, Su T, Ferrari R, Li B, Vatakis D, et al. (2011) Genome-wide binding map of the HIV-1 Tat protein to the human genome. PLoS One 6(11): e26894.
- Mazur PN, Rabalski L, Gromowski T, Grzegorz N, Michal K, et al. (2021) Expansion of a SARS-CoV-2 delta variant with an 872 nt deletion encompassing ORF7a, ORF7b and ORF8, Poland, July to August 2021. Euro Surveill 26(39): 2100902.
- Matsuoka K, Imahashi N, Ohno M, Ode H, Yoshihiro N, et al. (2022) SARS-CoV-2 accessory protein ORF8 is secreted extracellularly as a glycoprotein homodimer. J Biol Chem 298(3): 101724.
- McCarthy KR, Rennick LJ, Nambulli S, Robinson M, Bain WG, et al. (2021) Recurrent deletions in the SARS-CoV-2 spike glycoprotein drive antibody escape. Science 371(6534): 1139-1142.
- Meinberger D, Koch M, Roth A, Gabriele H, Jannik S, et al. (2021) Analysis of IgM, IgA, and IgG isotype antibodies directed against SARS-CoV-2 spike glycoprotein and ORF8 in the course of COVID-19. Sci Rep 11(1): 8920.
- Neches RY, Kyrpides NC, Ouzounis CA (2021) Atypical divergence of SARS-CoV-2 Orf8 from Orf7a within the coronavirus lineage suggests potential stealthy viral strategies in emmune evasion. mBio 12(1): e03014-20.
- Nemudryl A, Nemudraia A, Wiegand T (2021) SARS-CoV-2 genomic surveillance identifies naturally occurring truncation of ORF7a that limits immune suppression. Cell Reports 35(9): 109197.
- Ohki S, Imamura T, Higashimura Y, Matsumoto K, Mori M (2021) Similarities and differences in the conformational stability and reversibility of ORF8, an accessory protein of SARS-CoV-2, and its L84S variant. Biochem Biophys Res Commun 563: 92-97.
- Omoru OB, Pereira F, JangaSC, Manzourolajdad A (2022) A Putative long-range RNA-RNA interaction between ORF8 and Spike of SARS-CoV-2. PLoS One 17(9): e0260331.
- Parada CA, Roeder RG (1996) Enhanced processivity of RNA polymerase II triggered by Tat-induced phosphorylation of its carboxy-terminal domain. Nature 384: 375-378.
- Patarca R, Haseltine WA (2023) Intragenomic rearrangements involving 5′-untranslated region segments in SARS-CoV-2, other beta coronaviruses, and alphacoronaviruses. Virol J 20(1): 36.
- Pereira F (2020) Evolutionary dynamics of the SARS-CoV-2 ORF8 accessory gene. Infect Genet Evol 85: 104525.
- Raha T, Cheng SW, Green MR (2005) HIV-1 Tat stimulates transcription complex assembly through recruitment of TBP in the absence of TAFs. PLoS Biol 3(2): e44.
- Rashid F, Suleman M, Shah A, Dzakah EE, et al. (2021) Mutations in SARS-CoV-2 ORF8 altered the bonding network with interferon regulatory Factor 3 to evade host immune system. Front Microbiol 12: 703145.
- Sievers F, Wilm A, Dineen DG, Gibson TJ, Karplus K, et al. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Molecular Systems Biology 7: 539.
- Slanina, H, Madhugiri R, Bylapudi G, Karin S, Nadja K, et al. (2021) Coronavirus replication-transcription complex: Vital and selective NMPylation of a conserved site in nsp9 by the NiRAN-RdRp subunit. Proc Natl Acad Sci 118(6): e2022310118.
- Suharsono H, Mahardika BK, Sudipa PH, Tri K, Ida B, et al. (2023) Consensus insertion/deletions and amino acid variations of all coding and noncoding regions of the SARS-CoV-2 omicron clades, including the XBB and BQ.1 lineages. Arch Virol 168(6): 156.
- Su YC, Anderson DE, Young BE, Linster M, Zhu F, et al. (2020) Discovery and genomic characterization of a 382-Nucleotide Deletion in ORF7b and ORF8 during the early evolution of SARS-CoV-2. mBio 11(4): e01610-20.
- Takatsuki H, Fahmi M, Hamanishi K, Sakuratani T, Kubota Y, et al. (2022) In silico Analysis of SARS-CoV-2 ORF8-Binding Proteins Reveals the Involvement of ORF8 in Acquired-Immune and Innate-Immune Systems. Front Med (Lausanne) 9: 824622.
- Tao K, Tzou PL, Nouhin J, Gupta RK, Tulio d, et al. (2021) The biological and clinical significance of emerging SARS-CoV-2 variants. Nature Review Genetics 22(12): 757-773.
- Telwatte S, Martin HA, Marczak R, Parinaz F, Albert V, et al. (2022) Novel RT-ddPCR assays for measuring the levels of sub-genomic and genomic SARS-CoV-2 transcripts. Methods 201: 15-25.
- Urcuqui IS, Castaño ME, Hernandez V, Laurent, Kumar A (2006) Nuclear Factor 90, a cellular dsRNA binding protein inhibits the HIV Rev-export function. Retrovirology 3: 83.
- Valcarcel A, Bensussen A, Álvarez B, Díaz J (2021) Structural analysis of SARS-CoV-2 ORF8 protein: pathogenic and therapeutic implications. Front Genet 12: 693227.
- Vora SM, Fontana P, Mao T, Valerie L, Ying Z, et al. (2022) Targeting stem-loop 1 of the SARS-CoV-2 5′ UTR to suppress viral translation and Nsp1 evasion. Proc Nat Acad Sci 119(9): e2117198119.
- Wang Q, Iketani S, Li Z, Liu L, Guo Y, et al. (2023) Alarming antibody evasion properties of rising SARS-CoV-2 BQ and XBB subvariants. Cell 186(2): 279-286.e8.
- Wang X, Lam JY, Chen L, Au SW, Yuen KY, et al. (2021) Mining of linear B cell epitopes of SARS-CoV-2 ORF8 protein from COVID-19 patients. Emerg Microbes Infect 10(1): 1016-1023.
- Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, et al. (2018) SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46(W1): W296-W303.
- Wu X, Xia T, Shin W., Yu KM, Jung W, et al. (2022) Viral Mimicry of Interleukin-17A by SARS-CoV-2 ORF8. mBio 13(2): e00402-22.
- Yang Y, Jiang XT, Zhang T (2014) Evaluation of a hybrid approach using UBLAST and BLASTX for metagenomic sequences annotation of specific functional genes. PLoS One 9(10): e110947.
- Young, BE, Fong SW, Chan YH, Mak TM, Li Wei, et al. (2020) Effects of a major deletion in the SARS-CoV-2 genome on the severity of infection and the inflammatory response: an observational cohort study. Lancet 396(10251): 603-611.
- Yu T, Ling Q, Xu M, Wang N, Wang L, et al. (2022) ORF8 protein of SARS‐CoV‐2 reduces male fertility in mice. J Med Virol 94(9): 4193-4205.
- Zhang Y, Zhang J, Chen Y, Yaochang Y, Baijin X, et al. (2020) The ORF8 protein of SARS-CoV-2mediates immune evasion through potently downregulating MHC-I. BioRxiv 118(23): e2024202118.
- Zhang Y, Chen Y, Li Y, Huang F, Luo B, et al. (2021) The ORF8 protein of SARS-CoV-2 mediates immune evasion through down-regulating MHC-Ι. Proc Natl Acad Sci USA 118(23): e2024202118.
- Zhao J, Qiu J, Aryal S, Hackett JL, Wang J (2020) The RNA Architecture of the SARS-CoV-2 3'-Untranslated Region. Viruses 12(12): 1473.
- Zinzula L (2021) Lost in deletion: The enigmatic ORF8 protein of SARS-CoV-2. Biochem Biophys Res Commun 538: 116-124.
- Zhou Y, Zhang K, Yin X, Nie Q, Ma Y (2016) HIV-1 tat protein enhances expression and function of breast cancer resistance protein. AIDS Res Hum Retroviruses 32(1): 1-3.
© 2023, Asit Kumar Chakraborty . This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






