- Submissions

Full Text
Cohesive Journal of Microbiology & Infectious Disease
Exploration of Bioinformatics Approaches to Investigate DPP4 is a Promising Binding Receptor in SARS CoV-2
Saher Javaid1, Toheed Fatima2, Maryam Fatima3, Muhammad Yaqoob4, Imran Zafar5*, Kompal Fayyaz6, Quratul Ain7 and Waqas Yousaf8
1KAM school of life sciences, Forman Christian College Pakistan, Pakistan
2Department of Health informatics, COMSATS University Islamabad, Pakistan
3Department of Biotechnology, Virtual University of Pakistan, Pakistan
4Department of Life Sciences, ARID University, Pakistan
5Department of Bioinformatics and Computational Biology, Virtual University, Pakistan
6Department of National Centre for Bioinformatics, Quaid-I-Azam University Islamabad, Pakistan
7Department of Chemistry, Government College Women University Faisalabad, Pakistan
8Department of Botany, The University of Lahore, Pakistan
*Corresponding author: Imran Zafar, Department of Bioinformatics and Computational Biology, Virtual University Pakistan, Pakistan
Submission: March 09, 2023; Published: May 09, 2023

ISSN 2578-0190 Volume6 issues4
Abstract
The pandemic infection of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) has been rapidly increasing the number of patients afflicted with coronavirus disease 2019 (COVID-19) since December 2019. It has been reported that the entry receptor for SARS-CoV-2 is unequivocally Angiotensin- Converting Enzyme 2 (ACE2). However, it remains an open question whether ACE-related inhibitors or drugs can modify ACE2 activity and affect the viral activity and disease severity of SARS-CoV-2. This study was undertaken to determine the most promising binding receptor DPP4 with ACE2 to counteract its interaction with SARS-CoV-2 viral infection. To achieve this, Insilco studies were conducted based on retrieved data from various online databases. Molecular docking analyses have demonstrated that the spike protein Receptor Binding Domain (S-RBD) of SARS-CoV-2 efficiently binds to the DPP4 receptor, especially to the S-RBD vital residues of the RBP-binding motif located in the DPP4 receptor. Consequently, DPP4 may play a crucial role in elucidating the common pre- and post-COVID-19 symptoms of unknown etiology. This study also investigated the potential of epicatechin as a potent natural inhibitor that can be utilized to block or at least weaken the SARS-CoV-2 entry and its subsequent invasion. However, in vitro experiments are necessary to validate the effectiveness of epicatechin against the activity of the human ACE2 receptor.
Keywords: COVID-19; SARS-CoV-2; DPP4; ACE2; Spike protein; S-RBD
Introduction
DPP4 (also known as CD26) is a transmembrane protein that is expressed on the surface of various cells in the body, including immune cells, endothelial cells, and epithelial cells [1,2]. It plays a role in a variety of physiological processes, including immune regulation, glucose metabolism, and signal transduction. Recent studies Fani et al. [3] & Li et al. [4] have suggested that DPP4 may be involved in the pathogenesis of several viral infections, including MERS-CoV and SARS-CoV-2. These viruses have spike proteins on their surface that can interact with specific host cell receptors to facilitate entry into the cell. In the case of MERS-CoV, DPP4 has been identified as a functional receptor for the virus, and it is thought that the virus enters cells by binding to DPP4 on the cell surface [5].
In addition to its potential involvement in viral infection, DPP4 has been linked to a variety of other biological processes. For example, it is involved in glucose metabolism and insulin signaling, and has been implicated in the pathogenesis of type 2 diabetes [6]. DPP4 is also involved in the immune response, as it can cleave and inactivate several cytokines that are involved in regulating inflammation and immune function. There is also evidence to suggest that DPP4 may play a role in cancer progression, as it has been shown to be upregulated in several types of cancer and to promote tumor growth and metastasis [7]. DPP4 has been implicated in the pathogenesis of several other diseases, including asthma, liver disease, and cardiovascular disease. Because of its diverse roles in physiology and disease, DPP4 has been the target of numerous pharmacological interventions. Inhibitors of DPP4 are used as a treatment for type 2 diabetes, as they can increase insulin secretion and improve glucose control. DPP4 inhibitors are also being investigated for their potential use in other diseases, including cancer and inflammatory disorders [8].
DPP4 is an enzyme that cleaves peptides and proteins and has been shown to have a wide range of substrates [9]. In addition to its role in cleaving cytokines and incretins, it has been implicated in the processing of neuropeptides, chemokines, and growth factors. The role of DPP4 in inflammation is particularly interesting, as it has been shown to have both pro- and anti-inflammatory effects. DPP4 can promote inflammation by cleaving and inactivating anti-inflammatory cytokines such as IL-10. DPP4 can also have anti-inflammatory effects by cleaving and inactivating chemokines and other pro-inflammatory peptides. The net effect of DPP4 on inflammation is likely to depend on the specific context in which it is acting [10].
There is also evidence to suggest that DPP4 may play a role in the regulation of the Renin-Angiotensin System (RAS), which is involved in the regulation of blood pressure and fluid balance [11]. ACE2, which has been extensively studied as a receptor for SARS-CoV-2, is also involved in the RAS system, and there is some evidence to suggest that DPP4 may interact with the RAS system in a similar way [12].The role of DPP4 in physiology and disease is complex and multifaceted. While much research has focused on its role in glucose metabolism and diabetes, there is growing interest in its potential involvement in viral infections such as SARS-CoV-2, as well as in cancer, inflammation, and other diseases. Molecular docking and molecular dynamic simulation are computational modeling approaches Idris et al.[13] that were used to illustrate the most plausible pathway for Binding and stability in solvents.
Post dynamics analysis and, more specifically, rescoring based on computational methods, were used to minimize the prediction error in virtual screening. Utilizing molecular dynamics, an unfolded protein may be folded back into its physiologically folded state so that it can be leveraged in new therapeutic protocols. The position of the ligand within the active site can be ascertained using virtual screening results and docking data as per the investigation of earlier research [14]. To understand the molecular mechanism of action, molecular dynamics modeling is utilized to monitor the stable bond formation and molecule orientations concerning their catalytic residues [13].
To further understand their molecular mechanism of action, MD modeling is employed to monitor the stable bond formation and molecule orientation concerning their SARS-CoV S-RBD/ ACE2 complexes [15]. Along with these elements, a complete MD investigation was done to look at how the SARS-CoV-2 spike was binding to receptors and activating proteases. Retinol and retinoic acid are considered to be potentially effective treatments for COVID-19 infection and its unidentified etiological symptoms in the context of polymer dynamics and the pharmacological viewpoint on SARS-CoV-2 and their variants [16]. Along with these elements, a comprehensive MD study was carried out to understand polymer dynamics and the pharmaceutical viewpoint, both of which ultimately help the full investigations of development where Retinol Receptor-DPP4 as a new binding receptor in SARS CoV-2 pathogenesis [15].
Our aim is very diverse to explore the mechanistic process for the investigation of Retinol Receptor-DPP4 as a Novel Binding Receptor in SARS CoV-2 using some computational approaches where we strategize to anticipate the receptor usage infectivity of potential SARS-CoV to recognize their expected historical roots or genetic elements based on the patterns of their spike proteins and the known particle structural features of the initial SARSCoV S-RBD/ACE2 complexes. Here, based on the newly published data of the 2019-nCoV S-RBD, we consistently use this forecasting methodology to provide unprecedented insights into the receptor use and anticipated capacity to invade 2019-nCoV. Here, using the SARS-CoV spike as a baseline, we investigate the receptor binding & protease activations of the SARS-CoV-2 spike. Our findings, which are based on computational analysis, reveal important SARS-CoV-2 cell ingress pathways, where spike protein from the virus specifically binds to an integral receptor complex, probably assisting in signal transduction, cell transmissibility, and broad transmission (DPP4). We extensively examine the method by which DPP4, which recognizes RBP-retinol and triggers its release and internalization, facilitates Retinol (vitamin A) absorption into cells. The findings settle prior claims on SARS-CoV-2, where Retinol and retinoic acid are promising options for COVID-19 infection and its unknown etiological symptoms. Our Mechanistic Investigations are very surprising strategies where Novel Binding Receptor in SARS CoV-2 provides breakthrough feedback to regulate S-RBD/ ACE2 complexes in the context of Spike proteins to boost escaping immune regulation, that’s why the current investigation provides a novel insight into prospective treatment meansvia targeting SARSCoV- 2 specific sites.
Material and Methods
Sequence retrieval
The amino acid sequence of SARS CoV-2 spike protein with DPP4 and ACE2 (2ONC, 7EFP) for the Spike-ACE2 and DPP4 receptor was retrieved from the protein data bank in FASTA format as per earlier researchers Ahmad et al. [17] & Zafar et al. [18] to determine the 3-D structure of the target protein for further conformational investigations.
Molecular docking
HDOCK server (http://hdock.phys.hust.edu.cn/) was used to perform molecular docking to assess the binding mode of SARS CoV-2 spike protein with DPP4 and ACE2. PDB accession numbers (2ONC, 7EFP) for the Spike-ACE2 and DPP4 receptor proteins illustrate their binding mechanism. To evaluate whether the spike protein of the virus binds with the DPP4 receptor or not and, if so, with what affinity the binding happens, we studied the Spike- ACE 2 and DPP4 receptor proteins. The HDOCK server was used to conduct docking studies on the Spike-ACE 2 and DPP4 receptor proteins. The spike protein was put into the SAMSON software after downloading it from the PDB with accession number 6MOJ. Then, we separated the ACE2 and Spike proteins to conduct docking between them. Finally, we used the result as a control result to test and know how complex the spike-DPP4 receptor is and how much its efficiency, after that, the HDOCK server performed global docking to sample putative binding modes using an FFT-based search method. Following that, the putative binding modes were evaluated.
Protein-protein interaction network analysis
The DPP4 receptor protein was submitted to STRING v10.0 (https://string-db.org/) to analyze functional DPP4-associated networks, where interactions were analyzed at a medium confidence level as per the investigation of earlier researchers [19- 22].
Molecular dynamics simulation
The outcomes of simulations of molecular dynamics are immensely complex [23,24]. The Cartesian values of each atom in the system are recorded together with each time interval of the trajectory, which can again fluctuate in length from hundreds to millions of steps. More analysis is, therefore, necessary to extract the pertinent information from the data. The RMSD is a commonly used parameter for topological isolation across variables. It establishes the usual separation between a collection of atoms such as the backbone atoms of a protein [25]. The number serves as a gauge of how much the protein shape has altered when the RMSD is calculated over two different sets of atomic coordinates, such as two points in the trajectory. The RMSF determines the quantum state mean distances throughout time (for instance, from a peptide sequence) from a point of comparison (typically the timeaveraged position of the particle). Therefore, RMSF investigates the structural components that deviate most from their mean composition [26,27].The trajectory files were analyzed using the g_rms and g_rmsf GROMACS utilities to obtain the RMSD and Root Mean Square Fluctuation (RMSF) values. The numbers of distinct intermolecular hydrogen bonds formed during the simulation were calculated using the g_hbond utility. The trajectory files of the Principal Component Analysis (PCA) were analyzed using the g_ covar and g_anaeig utilities in GROMACS, in this order. The analysis of the secondary structure elements of the protein was performed using the do_dssp command, which utilizes the DSSP program [28].
Result and Discussion
We use computational approaches to investigate the mechanistic process for investigating the role of the retinal receptor-DPP4 as a novel binding receptor in SARS CoV-2. We plan to anticipate the receptor usage infectivity of potential SARS-CoV to identify their anticipated genetic or historical origins based on the patterns of their spike proteins and the known particle structural features of the initial SARS-CoV S-RBD/ACE2 complexes. Here, we regularly employ this forecasting approach based on the recently released data of the 2019-nCoV S-RBD to offer hitherto unheard-of insights into the receptor utilization and expected invading potential of 2019-nCoV.Furthermore, we study the receptor binding and protease activations of SARS-CoV-2 spike using SARS-CoV spike as a baseline. Our results, which are based on computational analysis, highlight significant SARS-CoV-2 cell entry routes, where the virus’s spike protein selectively attaches to with an integral receptor complex, likely facilitating signal transduction, cell transmissibility, and widespread transmission (DPP4).
Table 1:Complex Template Information of (DPP4 and ACE2).
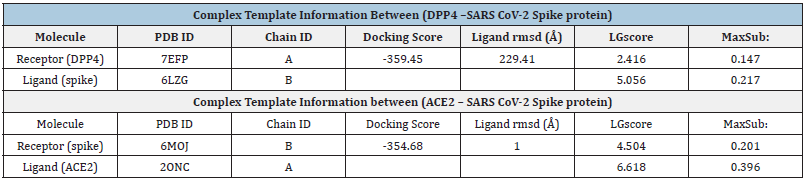
The intention was to learn more about the molecular mechanism as per earlier researchers Agarwal [29] & Dias et al. [30] through which the SARS-CoV-2 virus causes patients to have difficult symptoms by examining the binding affinity and mode of interaction between the two proteins. This might pave the way for therapeutic strategies that do not only rely on ACE2 to make first contact with the spike protein of the virus. In this work, the manner and process of the interaction were investigated by producing and docking the spike protein, ACE2 protein, and DPP4 receptor using the HDOCK server and our investigation was compared to earlier studies. We reinvestigate whether DPP4, particularly recognizes RBP-retinol and encourages its escape and enculturation, improves Retinol (vitamin A) absorption into cells. The findings contradict prior claims about SARS-CoV-2 that retinol and retinoids are efficient therapies for COVID-19 sickness and associated etiologically ambiguous symptoms. Because of our deterministic explorations, which are very remarkable methods where Novel Binding Receptor in SARS CoV-2 provides ground-breaking feedback to regulate S-RBD/ACE2 complexes in the context of Spike proteins to boost attempt to escape immunologic regulatory, the current study offers a novel insight of potential therapeutic strategies by targeting SARS-CoV-2 specific sites (Table 1).
Protein-protein docking (DPP4 and SARS CoV-2 spike protein)
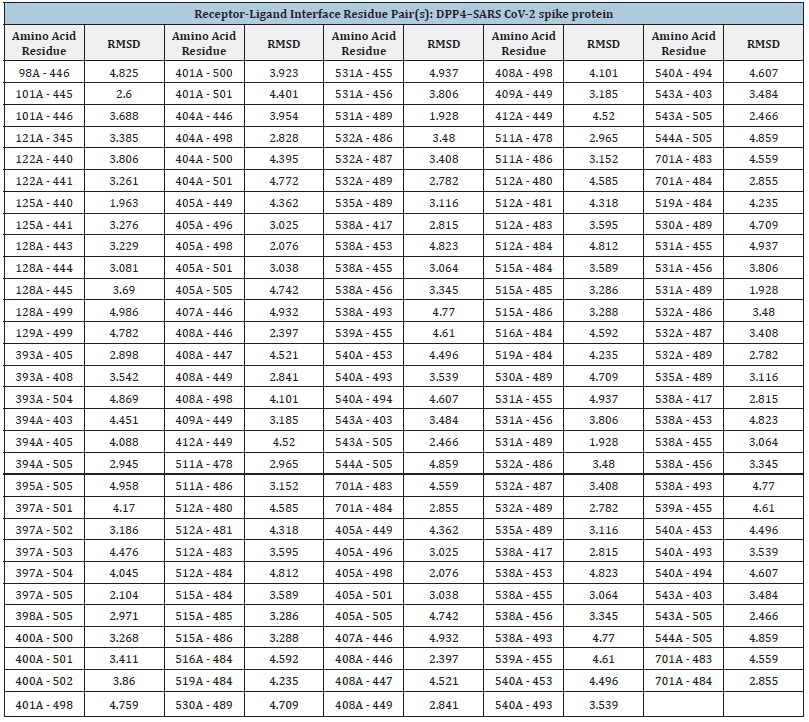
By using molecular docking, the molecules in libraries are compared based on their potential for the selected receptors as per the role of [30]. The folding of the biomolecules into the cavities of the receptor is necessary for the binding energy, which depends on a number of interactions, including charge transfer, hydrogen bonds, Van der Waals interactions, and others (Table 2). We observed that Hydrogen bonding can be ordinary or non-conventional, therefore the production of regular hydrogen bonds (H-bonds) between molecules with adequate functional groups that may serve as H-bond acceptors is dependable as per earlier research. The static phase of the receptor serves as the foundation for molecular docking screening, thus it’s critical to understand the MD trajectories across this interaction between receptor and ligands as well as the change in energy throughout dynamics. The interactive residues of the target protein DPP4 and the ligand spike protein can aid the design and development of more efficient and specific drugs for their target protein. The docking of DPP4 target protein with the viral spike protein revealed the involvement of the spike protein in the extracellular and membrane part of the DPP4 receptor. It showed the amino acid residues of DPP4 interacting with those of the spike protein, which are responsible for the protein-protein complex formation. The DPP4-spike protein complex (PDB ID 7EFP and 6LZG, respectively) reveals that chains A and B have Align lengths of 582 and 194, and then the quarry coverage of 0.793 and 1.000 and sequence identity of 96.2% and 100,0%, respectively. The surface view of the complex indicates that the binding pocket of the DPP4-spike and spike-ACE2 protein complexes have RMSD values of 189.44Å and 1.00Å, respectively. The docking scores are -341.21, and -354.68kcal/mol, and the quality of the ACE receptor and the ligand represented by the LG score and Max Sub scores are 2.416, 0.147, respectively, confirming the correctness of the structures. On the other hand, the DPP4 receptor protein’s LG score and Max Sub scores are 5.056, and 0.217, showing that the structures are very good and correct for the spike ligand and the receptor protein, as seen in (Table 1).
Table 2:DPP4 - SARS CoV-2 spike protein complex interaction data.
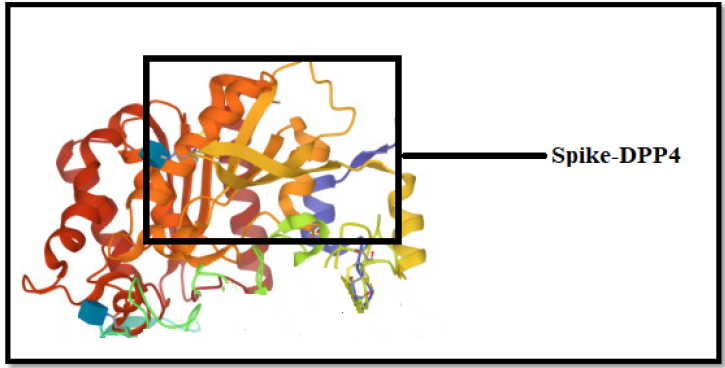
A virus’s Receptor-Binding Domain (S-RBD), which is part of its “spike” region and allows docking to host receptors for entrance into cells and invasion, is an integral part, as shown in (Figure 1). Here, we defined the Receptor-Binding Domain (S-RBD) of the SARS-CoV-2 S protein as observed that the S-RBD protein was tightly coupled to human DPP4 receptors. SARS-CoV-2 S-RBD attracted the DPP4 receptor considerably better than SARS-CoV S-RBD and was able to prevent activation. The S-RBD is a core part of viral spike glycoprotein or a crucial factor that allows virus infections to bind to various bodily receptors like the DPP4 receptors and readily penetrate to transmit illness is the attachment of the S-RBD to the spike domains. The RDB binding motif outside the cell, the DPP4 receptor’s membrane component outside the cell, and the receptor’s cytosolic third component are all shown in (Figure 1).
Figure 1:In this diagram, we depict three parts of the DPP4 receptor protein, where we indicate S-RBD, NTD, extracellular, intramembrane, and cytosol.
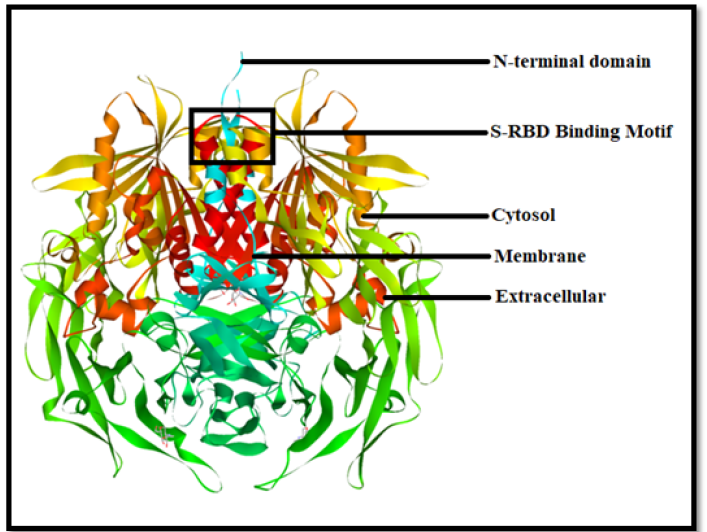
Substantial research has focused on SARS-CoV-2 spike protein changes in the S-RBD throughout this aspect. A minimal investigation has been carried out on deletions & mutations in the N-Terminal Domain (NTD), which lies close to the S-RBD. Many of these are found inside certain sheet-linking loops that are particularly long in SARS-CoV-2 compared to SARS-CoV or other related retroviruses. We demonstrate that the spike protein’s NTD loops in (Figure 1), which are both necessary for being recognized by a variety of autoantibodies, are maintained by both short- and long-contacts. Isolates demonstrating great infectiousness may have an impact on the data of NTD loops, similar to alterations in amino acids. In (Figure 1), we can see the cytosol area and extracellular membrane, where SARS spike protein is mostly concentrated in the lysosome and localized in the ER or ERGIC area, as per earlier research on intracellular trafficking and localization. The several new motifs found in the cytoplasm of the SARS spike protein may aid in the localization of the ER or ERGIC.
.The SARS-CoV-2 pattern interactions of spike protein are captured via the 2ONC receptor based on docking as shown in (Figure 2). The three alternate forms of the SARS-CoV-2 spike protein are discernible with the 2ONC receptor, which seems before and after the viral and cell envelopes unite. We observed that spikes bind to human cells via the 2ONC Receptor and then undergo a dramatic conformational shift in the context of the SARS CoV-2 Spike proteins in silico interactions. In the context of the 2ONC receptor, Spike proteins link their outer membranes to the protection of human cells by folding in on themselves like a jackknife and further it opens the door to investigate coronavirus infection.
Figure 2:Conformational Shift of SARS-CoV-2 Spike Proteins Upon Binding to the 2ONC Receptor.
Protein-protein interaction network/
Figure 3:Protein-protein interaction network of DPP4 receptor.
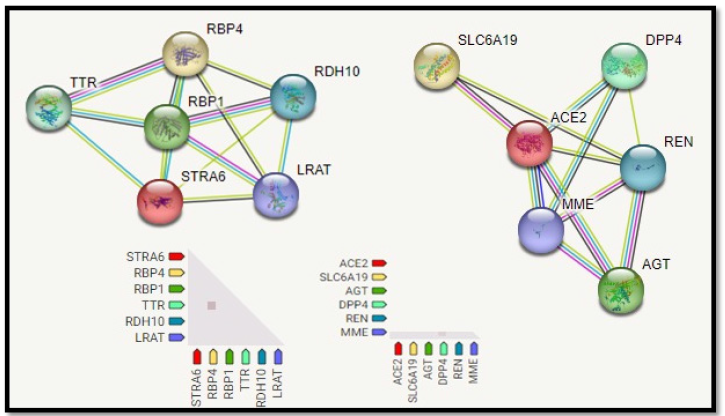
The Protein-Protein interaction associative network for the DPP4 receptor through STRING server shows that the active interaction sources were set based on seven parameters: experiments, co-expression, gene fusion, co-occurrence, databases, text mining, and neighborhood with a maximum of five interacting partners from both shells of interactions. The red color node describes query proteins, and the other colored nodes represent the first shell of interactors. The network interaction showed the following specifications number of nodes 7, number of edges 10, average node degree 2.86, local clustering coefficient 0.895, expected number of edges 6, and PPI enrichment p-value of 0.0973. The functional interactive network formed by the DPP4 receptor protein was analyzed at the medium confidence level (0.40), and it is shown in (Figure 3). Interactions between DPP4 and other proteins have been observed. These proteins include RBP4 Retinol-Binding Protein 4, which mediates retinol transport in blood plasma; TTR transthyretin, which transports thyroxine from the bloodstream to the brain; Retinol-Binding Protein 1, which accepts retinol from the transport protein DPP4, TTR, lRAT , RBP1, RDH10, and RBP4 which mediates the control of a large number of enzymes, ion channels, aquaporins, and other proteins.
Binding energy analysis of STRA-6 and SARS CoV-2 spike protein complex
The interactive residues of the target protein DPP4 and the ligand spike protein may help design and develop potential anti- SARS CoV-2 drugs that are more efficacious and specific for their biological targets. The docking of DPP4 target protein with SARS CoV-2 spike protein reveals the involvement and interaction of the spike protein into the extracellular and membrane part of the DPP4 receptor and amino acids residues of DPP4. The corresponding distances for the residue contacts between proteins DPP4 and the spike protein are listed in (Table 2).
Molecular Dynamics (MD) Simulation
In order to confirm the stability of the predicted docked ligand-ACE 2 and STRA 6 complexes, a study of all-atom Molecular Dynamics (MD) simulations was carried out. Such a study would be helpful to get further understanding of the dynamic interactions between the ligand and the protein receptors 2ONC and 7EFP, as well as to assess the ligand’s major binding contacts with important enzymatic domain regions. Therefore, a 100ns all-atom MD simulation took into consideration the expected ligand-protein interactions for both the ACE 2 and STRA 6 proteins. We looked at the dynamic behavior of the ACE 2 (2ONC) and STRA 6 (7EFP) receptor proteins in complex with SARS CoV-2 spike protein as a complex as a consequence of the docking study results mentioned above. Calculations were made for the Root Mean Square Deviation (RMSD) and Root Mean Square Fluctuation (RMSF).The primary goal of the MD simulation studies was to investigate the protein’s positional and structural changes. Root Mean Square Fluctuation is a useful method for detecting regional changes throughout the chain of interactions involving a protein.
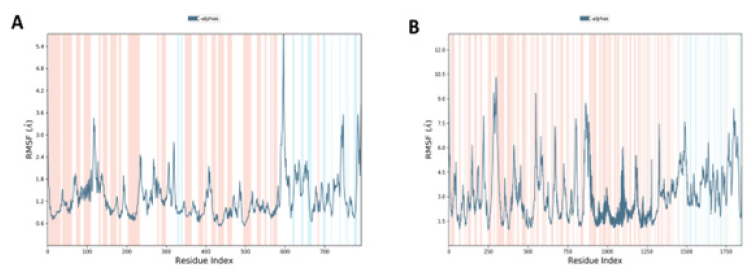
MD studies revealed the RMSF plot in (Figure 4) for ACE 2-SARS CoV-2 spike protein complex, and DPP4-SARS CoV-2 spike protein complex, respectively. An MD modeling investigation showed that simply changing the conformation of the N- terminal site in the range of 580:800, ACE 2 may successfully activate the biological pathway. The RMSF of the ACE 2 receptor protein in association with SARS CoV-2 spike protein was assessed, and the results showed that the protein has a high RMSF in the bound state that ranges from 0.6 to 5.6.The variations in RMSF suggest that the protein’s structure has changed as a result of the binding of its residues to the ligand. The involvement of these residues in the interaction with the ligand is shown by a large variation in RMSF at the N-terminal locations. The residue position 690 had the most variation. The outcomes, therefore, demonstrate an active interaction between the ligand and the target leading to protein conformational changes. DPP4 activates at the C-terminal in the range of 250-500 and then in the middle in the range of 500-1000 with significant RMSF ranging from 1.3-10.5. The residue position 270 had the most variation.
Figure 4:Analysis of RMSF of the carbon alpha for 6A (ACE2– SARS CoV-2 spike protein complex) and 4B (DPP4 – SARS CoV-2 spike protein complex).
During the MD simulations, RMSD was utilized to assess the stabilities of the ACE 2 and DPP4 complex with the SARS CoV- 2 spike protein for 100ns for each trajectory frame. The protein complexes’ time (ns) for frame x was calculated with respect to the original structures along the 100 ns trajectory using the provided trajectories. RMSD for ACE 2 was found to be 3.4 and 10.6 for DPP4 (Figure 5). The SSE composition for each trajectory frame over the simulation (i.e., 100ns) was determined and shown in (Figure 5). For the ACE2 complex, the α-helix and β-strand accounted for 46.98% of the total SSE, whereas the DPP4 complex was 45.38% of the total SSE.
Figure 5:RMSD analysis of the 7A (ACE2– SARS CoV-2 spike protein complex) and 7B (DPP4 – SARS CoV-2 spike protein complex).
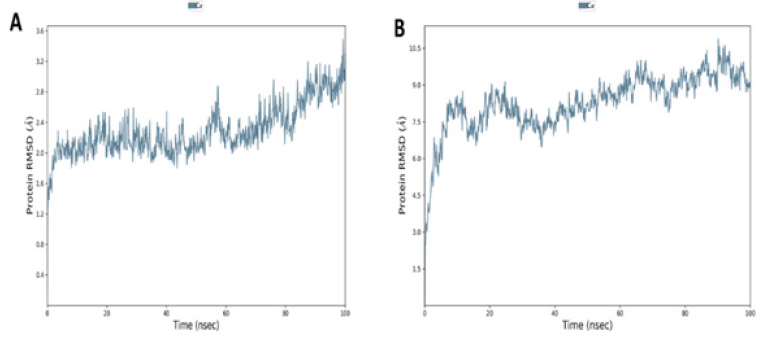
Figure 6:Characterizing Protein Structure Dynamics of ACE2 and DPP4 Receptors upon Binding to SARS-CoV-2 Spike Protein Using SSE Analysis.
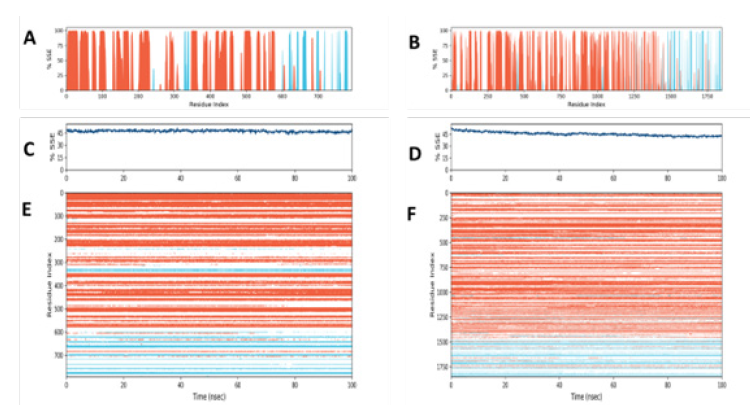
In this study, we monitored the distribution of Secondary Structure Elements (SSEs) such as α-helices and β-strands throughout the simulation of ACE2 and DPP4 receptors binding to SARS-CoV-2 spike protein. (Figures 6A & 6B) illustrate the distribution of SSEs by residue index throughout the protein structure, while Figures 6C & 6D summarize the composition of SSEs for each trajectory frame over the 100 ns simulation. Moreover, we tracked the SSE assignment of each residue over time for ACE2-SARS CoV-2 spike protein (Figure 6E) and DPP4- SARS CoV-2 spike protein (Figure 6F), where red and blue colors represent the SSE assignment of α-helix and β-strand, respectively. Our findings provide valuable insights into the protein structure dynamics of ACE2 and DPP4 receptors upon binding to SARS-CoV-2 spike protein, which could aid in the development of effective therapeutics against COVID-19.Conclusion
The objective of our study was to explore the potential role of DPP4 as a binding receptor in SARS-CoV-2 pathogenesis, in addition to investigating the therapeutic potential of targeting DPP4 in COVID-19 treatment. Our findings provide a promising avenue for future research in the field of COVID-19 treatment. Further exploration of DPP4 as a binding receptor for SARS-CoV-2 can shed light on the pathogenesis of the virus and contribute to the development of effective treatments. Additionally, the investigation of DPP4 as a potential therapy for COVID-19 warrants further clinical trials to evaluate its safety and efficacy. Further research can also investigate the molecular mechanisms behind the observed binding affinity between DPP4 and SARS-CoV-2 Spike protein, which can contribute to the development of novel therapeutics targeting this interaction. Remarkably, our study demonstrated that SARS-CoV-2 Spike protein exhibited a high docking score with human DPP4 with low binding energy. The docking score of the SARS-CoV-2 spike protein was stronger than the docking score of the spike protein with ACE2. Based on our findings, targeting DPP4 may be a promising and effective treatment strategy for COVID-19 infection and its unknown etiological symptoms. It is worth mentioning that further investigations are needed to evaluate the safety and efficacy of DPP4-targeted therapies in clinical trials. Our study provides valuable insights into the potential of targeting DPP4 as a novel therapeutic strategy for COVID-19 and highlights the need for further research in this area.
References
- Matteucci E, Giampietro O (2009) Dipeptidyl peptidase-4 (CD26): Knowing the function before inhibiting the enzyme. Current Medicinal Chemistry 16(23): 2943-2951.
- Solerte SB, Di Sabatino A, Galli M, Fiorina P (2020) Dipeptidyl Peptidase-4 (DPP4) inhibition in COVID-19. Acta Diabetologica 57(7): 779-783.
- Fani M, Teimoori A, Ghafari S (2020) Comparison of the COVID-2019 (SARS-CoV-2) pathogenesis with SARS-CoV and MERS-CoV infections. Future Virology 15(5): 317-323.
- Li Y, Zhang Z, Yang L, Lian X, Xie Y, et al. (2020) The MERS-CoV receptor DPP4 as a candidate binding target of the SARS-CoV-2 spike. Iscience 23(8): 101400.
- Bassendine MF, Bridge SH, McCaughan GW, Gorrell MD (2020) COVID‐19 and comorbidities: A role for dipeptidyl peptidase 4 (DPP4) in disease severity? Journal of Diabetes 12(9): 649-658.
- Deacon CF (2019) Physiology and pharmacology of DPP-4 in glucose homeostasis and the treatment of type 2 diabetes. Frontiers in Endocrinology 10: 80.
- Wesley UV, Tiwari S, Houghton AN (2004) Role for dipeptidyl peptidase IV in tumor suppression of human non small cell lung carcinoma cells. International Journal of Cancer 109(6): 855-866.
- Yazbeck R, Howarth GS, Abbott CA (2009) Dipeptidyl peptidase inhibitors, an emerging drug class for inflammatory disease? Trends in Pharmacological Sciences 30(11): 600-607.
- Aertgeerts K, Ye S, Tennant MG, Kraus ML, Rogers J, et al. (2004) Crystal structure of human dipeptidyl peptidase IV in complex with a decapeptide reveals details on substrate specificity and tetrahedral intermediate formation. Protein Science 13(2): 412-421.
- Sedo A, Duke JS, Balaziova E, Sedova LR (2005) Dipeptidyl peptidase IV activity and/or structure homologs: Contributing factors in the pathogenesis of rheumatoid arthritis? Arthritis Research & Therapy 7(6): 253-269.
- Yaribeygi H, Maleki M, Sathyapalan T, Jamialahmadi T, Sahebkar A (2020) Incretin-based therapies and renin-angiotensin system: Looking for new therapeutic potentials in the diabetic milieu. Life Sciences 256: 117916.
- Badawi S, Ali BR (2021) ACE2 Nascence, trafficking, and SARS-CoV-2 pathogenesis: The saga continues. Human Genomics 15(1): 1-14.
- Idris MO, Yekeen AA, Alakanse OS, Durojaye OA (2021) Computer-aided screening for potential TMPRSS2 inhibitors: A combination of pharmacophore modeling, molecular docking and molecular dynamics simulation approaches. Journal of Biomolecular Structure and Dynamics 39(15): 5638-5656.
- Rather MA, Dutta S, Guttula PK, Dhandare BC, Yusufzai S, et al. (2020) Structural analysis, molecular docking and molecular dynamics simulations of G-protein-coupled receptor (kisspeptin) in fish. Journal of Biomolecular Structure and Dynamics 38(8): 2422-2439.
- Kalathiya U, Padariya M, Mayordomo M, Lisowska M, Nicholson J, et al. (2020) Highly conserved homotrimer cavity formed by the SARS-CoV-2 spike glycoprotein: A novel binding site. Journal of Clinical Medicine 9(5): 1473.
- Agarwal D, Zafar I, Ahmad SU, Kumar S, Sundaray JK, et al. (2022) Structural genomic information and computational analysis of emerging coronavirus (SARS-CoV-2). Bulletin of the National Research Centre 46(1): 1-16.
- Ahmad SU, Kiani BH, Abrar M, Jan Z, Ali Y, et al. (2022) A comprehensive genomic study, mutation screening, phylogenetic and statistical analysis of SARS-CoV-2 and its variant omicron among different countries. Journal of Infection and Public Health 15(8): 878-891.
- Zafar I, Iftikhar R, Ahmad SU, Rather MA (2021) Genome wide identification, phylogeny, and synteny analysis of sox gene family in common carp (Cyprinus carpio). Biotechnology Reports 30: e00607.
- Rather MA, Dhandare BC (2019) Genome-Wide identification of doublesex and Mab-3-Related transcription factor (DMRT) genes in nile tilapia (Oreochromis niloticus). Biotechnology reports 24: e00398.
- Zafar I, Pervez MT, Rather MA, Babar ME, Raza MA, et al. (2020) Genome-Wide identification and expression analysis of PPOs and POX gene families in the selected plant species. Biosciences Biotechnology Research Asia 17(2): 301-318.
- Karplus M, Petsko GA (1990) Molecular dynamics simulations in biology. Nature 347(6294): 631-639.
- Hornak V, Okur A, Rizzo RC, Simmerling C (2006) HIV-1 protease flaps spontaneously open and reclose in molecular dynamics simulations. Proceedings of the National Academy of Sciences 103(4): 915-920.
- Henzler-Wildman K, Kern D (2007) Dynamic personalities of proteins. Nature 450(7172): 964-972.
- Lindahl ER (2008) Molecular dynamics simulations. Molecular Modeling of Proteins, Springer, New York, USA, pp. 3-23.
- Agarwal S, Mehrotra R (2016) An overview of molecular docking. JSM Chem 4(2): 1024-1028.
- Dias R, de Azevedo J, Walter F (2008) Molecular docking algorithms. Current Drug Targets 9(12): 1040-1047.
- Elkazzaz M, Ahmed AKK, Shamkh IM, Abo El Magd M (2021) STRA6 (vitamin A receptor), as a Novel binding receptor of COVID-19. Science Open Preprints.
- Elkazzaz MR, Haydara T, Abo AE, Sahyon H, Shamkh IM, et al. (2021) Discovery of the retinol STRA6 receptor as a novel binding receptor for SARS-CoV-2 in COVID-19: In Silico Research.
- Kitakaze T, Yoshikawa M, Kobayashi Y, Kimura N, Goshima N, et al. (2020) Extracellular transglutaminase 2 induces myotube hypertrophy through G protein-coupled receptor 56. Biochim Biophys Acta Mol Cell Res 1867(2): 118563.
- Hunt PA, Ashworth CR, Matthews RP (2015) Hydrogen bonding in ionic liquids. Chemical Society Reviews 44(5): 1257-1288.
© 2023, Sávio Benvindo Ferreira. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






