- Submissions

Full Text
Cohesive Journal of Microbiology & Infectious Disease
A Brief Review of Antibiotic Resistance Among Pathogenically Important Bacteria
Aqib Saeed1, Fatima Ahsan1, Muhammad Nawaz1*, Chengcheng Liu2, Saadia Ijaz3 and Muhammad Saad khilji4,5,6
1Institute of Microbiology, Lahore
2Department of Pathogenic biology and Immunology, China
3Shahlimar Medical and Dental College, Lahore
4Department of Physiology, Lahore
5Laboratory of Immuno-Endocrinology, Denmark
6Department of Biochemistry and Molecular Biology, Australia
*Corresponding author: Muhammad Nawaz, Institute of Microbiology, Lahore, Pakistan
Submission: September 08, 2020; Published: September 23, 2020

ISSN 2578-0190 Volume4 issues2
Abstract
Antibiotics are known as “lifesaving drugs” and use for the treatment of infectious diseases. The use of antibiotics is not limited with the treatment of infectious diseases but also use prophylactically in other industries such as in livestock and agriculture. Unfortunately, due to extensive use of antibiotics, microbes develop resistance against antibiotics. Aim of the current review is to explore the history, causes, mechanisms of antibiotic resistance and alternative to antibiotics by examining the available literature. Antibiotic resistance is rising at an alarming rate and a major threat to the public health. A significant association was found in the antibiotic resistant infections with the level of antibiotic consumption. Inappropriate prescription, lack of awareness among the people and excessive use in the agriculture and livestock sectors are the main causes which are responsible for the resistance of microbes to antibiotics. Various mechanisms are involved in the antibiotic resistance such as mutation in genes, horizontal genes transfer, reduced permeability, alterations in target sites and enzymatic degradation. Furthermore, alternate options i.e. Photodynamic antimicrobial therapy, probiotics, medicinal plants and nanoparticles etc. can be used in health care setups to combat the increasing antibiotic resistant infections.
Introduction
Antibiotics, either cytotoxic or cytostatic, are used to treat infections. The mode of action of antibiotics could be either by inhibition of the synthesis of cell wall, proteins, deoxyribonucleic acid (DNA) or ribonucleic acid (RNA). It could be achieved by a membrane disorganizing agent or other specific actions [1]. Undoubtedly, antibiotics are a blessing to humans for the treatment of infections or microbes [2]. Various antibiotics have been used for the treatment purposes for a long time. During the mid-20th century antibiotics were considered as “Wonder drug”. The beginning of modern “Antibiotic era” was synonymously associated with two persons Alexander Fleming and Paul Ehrlich. The concept of antibiotics was like a magic bullet that selectively targets the microbes responsible for the diseases but at the same time would not affect the host [3]. The period from 1950 to 1970 was considered as a golden era for the discovery of novel antibiotic classes [4].
Undeniably, antibiotics have become one of the most important medical interventions needed for the development of complex medical approaches such as cutting-edge surgical procedures, solid organ transplantation and management of patients with cancer among others. Unfortunately, the sudden rise in antimicrobial resistance among the common bacterial pathogens is an alarming condition for the therapeutic accomplishment and it is also a potential threat to the critically ill patients. In fact, the World Health Organization (WHO) has categorized the antibiotic resistance as one of the three major threat to the public health in 21st century [5]. Antimicrobial resistance (AMR) is increasing globally presenting significant challenges to the prevention and treatment of common bacterial infections [6,7]. The possible consequences of this resistance are worse, the health costs also increased [8] and an increased risk of morbidity and mortality [9].
The European Center for Disease Control (ECDC) and Center for Disease Control and Prevention (CDC) Atlanta described the terminologies Multidrug resistant (MDR), Extensive drug resistant (XDR) and Pan drug resistant (PDR) bacteria [10]. According to them organism resistant to at least one agent in three or more antimicrobial agents is known as multidrug resistant (MDR), whereas non-susceptibility to at least one agent in all but susceptible to two or more antimicrobial agents are known as extensive drug resistant (XDR) and organisms resistant to all agents in all antimicrobial classes are defined as pan drug resistant (PDR) [11]. The treatment of infections caused by multi drug resistant (MDR) gram positive and gram-negative bacteria by conventional antibiotics seems to be difficult now. The poor infection control practices result in the dramatic rise in the antibiotic resistance that also spread in other patients and also in the environment [12]. Resistance of bacterial pathogens to the common antimicrobial therapies and emergence of multi drug resistant (MDR) pathogens are increasing at alarming rate. There are various challenges in the treatment of infections such as shortage of effective drugs, lack of successful preventive measures and availability of only few antibiotics in clinical pipelines. For effective treatment there is a need of development of novel treatment options and alternative antimicrobial therapies [13]. The current reviews focus on the mechanisms of antibiotic resistance, its origin, possible causes, and the alternatives other than antibiotics that will be helpful in combating the infections caused by the MDR and XDR bacteria in future.
History of Antibiotic Resistance
In 1940s antibiotics were first prescribed for the treatment of serious infections [11]. During World War II Penicillin was considered as the most potent drug for the treatment of bacterial infections [14]. However, shortly thereafter by the 1950s Penicillin resistance became a major clinical issue and many of the advances of the prior decade were threatened [15]. In order to combat this issue new beta-lactams antibiotics were discovered, industrialized and distributed [14,15]. Antibiotic resistance can occur as a natural selection process where nature empower all bacteria with some degree of low-level resistance [2]. A study from Thailand revealed that antibiotics such as sulfamethoxazole and trimethoprim (TMP-SMZ), ampicillin and tetracycline that were previously used in the treatment of non-cholera diarrhea disease have no longer any role in the treatment, nowadays, due to development of resistance against these antibiotics [16]. Within six years of the production of aminoglycosides, the aminoglycoside resistant Staphylococcus aureus strains were developed [17]. Methicillin was the first semisynthetic penicillinase-resistant penicillin that was used for the treatment of penicillinase producing Staphylococcus aureus strains. However, after starting its use in therapeutics methicillin resistance was reported very soon [18].
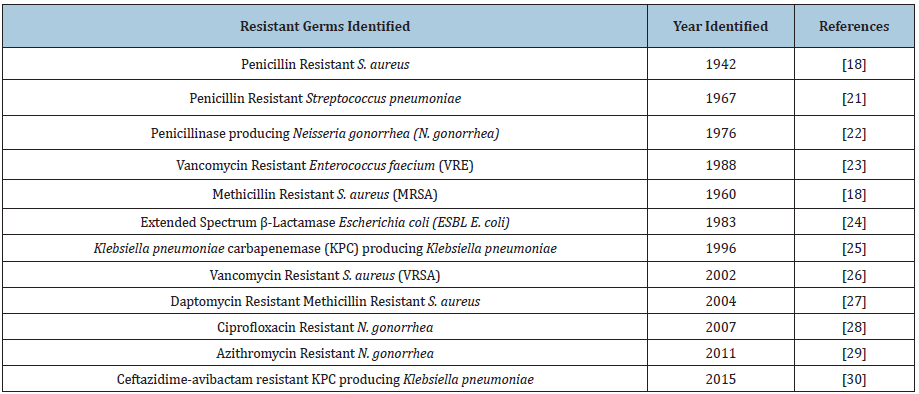
The first case of Methicillin Resistant Staphylococcus aureus (MRSA) was identified in United Kingdom in 1962 and later in United states in 1968 [14]. Moreover, in 1980s fluroquinolones were used initially to treat the gram-negative bacterial infections but later it was revealed that fluroquinolones were also effective for the treatment of gram-positive infections [19]. Later, quinolone resistance developed as stepwise attainment of chromosomal mutation particularly among the MRSA strains. Furthermore after 44 years of introduction of vancomycin in the market, the clinical isolates of Vancomycin Resistant Staphylococcus aureus (VRSA) were reported in 2002 [20-31] as shown in the (Table 1).
Table 1: Emergence of antibiotic resistant pathogens.
Antibiotics used in the agriculture are almost same as used in the clinical side and frequent use of these antibiotics leads towards the drug resistance [31]. The food chain can play a significant role in the transmission of antibiotic resistance from animals to human populations [32]. In developing countries animals receive antibiotics by food, water and parenterally. The exposure of microbes to antibiotics may develop resistance against these antibiotics [31]. The use of antibiotics in poultry and cattle feed as growth promoter increase the antibiotic resistance [2]. A study from Barcelona revealed that poultry might be a possible source of quinolone resistant E. coli in rural villages. The study also revealed that one-fourth of the children were found fecal carriers of quinolone resistant E. coli. Although these children were never exposed to the quinolones [33].
Causes of Antibiotic Resistance
Misuse
Various epidemiological studies have shown a direct correlation between the usage of antibiotics and rise in resistant bacterial strains [6]. In bacteria, genes can be transferred from one species to another species by the aid of mobile genetic elements such as plasmids. The horizontal gene transfer (HGT) can allow antibiotic resistance to be transferred among different species of bacteria [34]. Despite various cautions regarding the misuse, antibiotics are overprescribed globally. In several countries’ antibiotics are unregulated and available under the table without any prescription [6]. The outcomes of absence of guidelines are very easy and cheap access to the antibiotics which enhance the overuse of antibiotics. The easy online access to purchase the antibiotics without any prescription is also another example of misuse of antibiotics [35]. Various studies demonstrated that the old aged people such as 65 years or above have more chances to acquire bacterial infection as compared to the younger ones hence as a result old aged people consumes more antibiotics [36-38]. Furthermore due to presence of atypical signs such as fall, delirium and generalized weakness the diagnosis of bacterial infection is difficult in older aged people which in turn contributes to the overconsumption of antibiotics in the context of bacterial colonization or non-infectious diseases [39]. The misuse of antibiotics by physicians in intensive care units (ICUs) is also very common that is becoming a breeding place for the development and dissemination of antibiotic resistance bacteria [40]. The inappropriate use of antibiotics is prevalent globally, but the developing countries are mostly affected due to higher infection rates and limited resources [41,42]. Pakistan is a developing country which has a sub optimal health care set-up where antibiotics are sale over the counter [43,44]. The very limited financial resources, poor literacy rates and compromised health care management showed that the surveillance of health care set ups might not be takes place properly, therefore irrational use of antibiotics also takes place continuously [43]. Among the low-income countries Pakistan is classified as the 3rd highest country in the consumption of antibiotics. A rapid increase in the consumption of antibiotic is seen between 2000 and 2015 where antibiotic consumption increased from 0.8 to 1.3 billion (Up to 65%) defined daily doses (DDDs) [45].
Inappropriate prescription
Inappropriate prescription also plays an important role in the raise of antibiotic resistance. Various studies revealed that in 30-50% cases the selection of antibiotic agents, duration of antibiotic therapy is inappropriate [11]. Inaccurately advised antibiotics usually have uncertain therapeutic benefits and expose patients to potential complications of antibiotic therapy [46]. The subinhibitory and subtherapeutic concentration of antibiotics can play a vital role in the development of antibiotic resistance by genetic modification, for instance, by changes in the gene expression, by horizontal gene transfer (HGT) and by mutagenesis. The modification in the antibiotic resistant genes can increase the virulence of microorganisms whereas the alteration due to HGT and mutagenesis develop antibiotic resistance and its spread in the environment. Low level of antibiotics has shown the strain diversification in organisms such as Pseudomonas aeruginosa. While on the other hand the subinhibitory concentration of Piperacillin/ Tazobactam has shown various proteomic alterations in the Bacteroides fragilis [47]. In developing countries due to lack of qualified and well-trained staff, the poor trained or self-trained quacks pretend themselves as medical practitioners in rural areas [48].
Most of the practitioners in the rural areas are semi-literate and they don’t have any idea about the proper preservation of antibiotics, the harmful effects of antibiotics and inappropriate prescription of antibiotics they seem to have an antibiotic for every human ailment [49]. For instance, a study from Thailand revealed that a pharmacy technician advises rifampicin for urethritis and tetracycline for young children [50]. A survey of World Health Organization (WHO) revealed that there are only 0.97 physicians per 1000 people in Pakistan, as compared with India 0.70, in China 1.6 and in United states there are 2.56 physicians per 1000 people [51]. In Pakistan the resistance of bacteria towards the antibiotics is a serious issue. The analysis of antibiotics may be helpful to avoid the inappropriate antibiotic prescription that decrease the chances of resistance [52]. However, a study from southern Punjab, region of Pakistan described that majority of antibiotics prescribed without culture and sensitivity testing [41].
Excessive use of antibiotics in livestock and agriculture
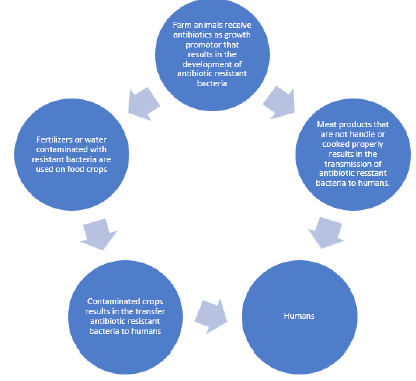
In livestock sector antibiotics are used as a source of growth promoter in both developing and developed countries [6,11,53]. In United States an estimated 80% of antibiotics are sold only for the growth, enhancement and protection of animals from the infections [15,53,54]. The treatment of animals with the antibiotics is considered as a best way to protect the animals from the infections, for their better health, and also for the improved yield quality [35]. Antibiotics use in livestock are ingested by humans when they consume food [55]. The transmission of antibiotic resistant bacteria from animals to humans was noticed first time 35 years ago [53]. Recently with the aid of molecular methods it is revealed that the antibiotic resistant bacteria transmit from farm animals to consumers by the meat products [53]. The spread of antibiotic resistant bacteria occurs in the following steps. Firstly, antibiotics used in the food of animals suppress or destroy the beneficial microbes and only those microbes survive that are resistant to antibiotics. In the second step these resistant bacteria transmit from animals to humans by food chain. After their transmission these bacteria cause serious infections in humans [11] as shown in the Figure 1. The use of antibiotics in the agriculture also disturb the environmental microbiome [11]. Upto 90% of antibiotics given to the livestock are excreted in the urine and stool, then widely spread through fertilizers, ground water and surface run off [53]. Furthermore, in western and southern US the streptomycin and tetracycline are used as pesticides. This practice also contributes to the exposure of microorganisms in the environment to growth inhibiting agents, altering the environmental ecology by increasing the proportion of resistant versus susceptible microorganisms [55].
Figure 1: Transmission of antibiotic resistant bacteria to humans by food chain.
Antibiotic Resistant Bacterial Infections
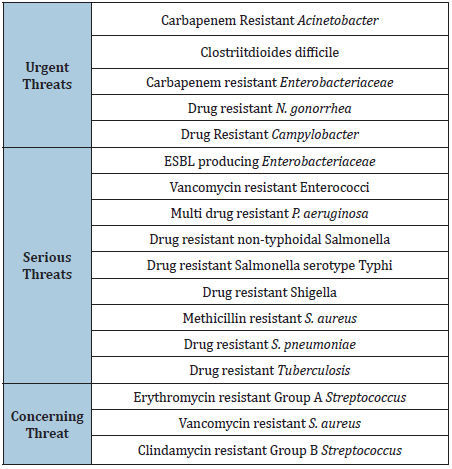
Infections due to antibiotic resistant bacteria are spreading very rapidly all over the world [55]. Various public health organizations have explained such speedy emergence of resistant bacteria as “disaster” or “frightening situation” whose consequences could be very devastating [47]. A report from Center for Disease Control and prevention (CDC) in 2013 declared that humans has reached now in the “Post antibiotic era” whereas in 2014 World Health Organization (WHO) cautioned that the antibiotic resistance crisis is now becoming dire [35]. Among gram positive pathogens Staphylococcus aureus and Enterococcus species currently pose the major challenges in terms of antibiotic resistance [11,56]. On the other hand, resistance in the gram-negative pathogens is also alarming because they are becoming resistant to almost all available antibiotics thus pushing us back in the “pre-antibiotic era” [11,56]. The center for diseases control and prevention (CDC) evaluated the antibiotic resistant infections on the basis of several factors such as clinical impact, economic impact, incidence, 10-year projection of incidence, transmissibility, availability of effective antibiotic, and barrier to prevention. In this evaluation CDC categorized the antibiotic resistant bacteria into 3 categories as “Urgent”, “Serious”, “Concerning” [11] as shown in the (Table 2).
Table 2: Classification of antibiotic resistant bacterial infections.
Impact of Antibiotic Resistance on Global Economy
The one of the biggest threats to the public health is antibiotic resistance that endanger the achievements towards the millennium development goals as well as the sustainable development goals [57]. ABR is also associated with higher morbidity, mortality, prolonged stay at hospitals and it also reduce the labor efficiency [58] The assessment of economic burden due to antibiotic resistance is still a gigantic challenge globally. The Hospital acquired infections (HAIs) only in United states cause 99,000 deaths a year. In 2006 only 50,000 deaths in United States occurred because of two HAIs, namely pneumonia and bacteremia that cost approximately $8 billion to the US economy [59]. The patients with antibiotic resistant bacterial infections need to stay at hospital for at least 13 days. Due to prolonged stay at hospital the cost for the treatment also increased. An estimated cost of $29,000 per patient reported for the treatment of antibiotic resistant bacterial infections. In total the economic losses of about $20 billion have been reported in the US whereas the losses of around $35 billion also reported in terms of loss of productivity due to antibiotic resistance in health care system [60].
Antibiotic Resistance Mechanism
Genetic mechanism
Bacteria have genetic diversity that make them able to respond to the various types of environmental threats that also include the presence of antibiotic molecule which may threatens their existence. Evolutionary perception revealed that bacteria utilize two major genetic approaches to acquire the antibiotic resistance. Among these genetic approaches one is mutation in genes and the second is acquisition of foreign DNA by horizontal gene transfer (HGT) [61].
Horizontal gene transfer (HGT): Bacteria acquire foreign DNA materials by horizontal gene transfer (HGT). It is one of the most important drivers of bacterial evolution and also responsible for the development of antibiotic resistance. The antimicrobial compounds used in the clinical settings mostly obtained from the natural environment (from soil). The bacteria acquire the external genetic material by three main strategies. By transformation, transduction and conjugation. The simplest type of horizontal gene transfer is transformation. But only a few clinically relevant bacterial species are able to naturally insert the naked DNA to develop resistance. In hospital environment the resistance is often developed by the process of conjugation. This method is very efficient for the gene transfer that involve the cell to cell contact and is likely to occur at high rates in the gastrointestinal tracts of humans under antibiotic treatment. In conjugation the mobile genetic elements are used as vehicle to share the valuable genetic information, although the direct transfer from chromosome to chromosome has also been well characterized [62].
Mutation in genes: In this mechanism a colony of bacterial cells isolated from a susceptible environment develop changes in their genes that disturb the functioning of antibiotic, results in the survival of organism in the presence of antibiotic molecule. Once development of resistant mutants take place, antibiotic only eradicate the susceptible population and the resistant population prevails. The mutation results in the antibiotic resistance, modify the working action of antibiotic by one of the following mechanisms. (1) By modifying the antimicrobial agent, (2) Stimulation of efflux pump mechanism to exclude out the harmful molecules, (3) Decline in the drug uptake and (4) Global changes in important metabolic pathways via modulation of regulatory networks [61].
Efflux pump stimulation: The membrane proteins that export the antibiotic molecules out of the cell and as a result maintain the low intracellular concentration is known as efflux pump [63]. As antibiotic molecules enter the cell, the efflux mechanism, at the same time, expels out these antibiotic molecules without letting them reach the target site [64]. These pumps are in the cytoplasmic membranes as compared to the porins that are located in the outer membrane [65]. There are 5 major families of efflux pump mechanism. These families differ in terms of structural conformation, energy source, range of substrate and these are widely distributed among different types of bacterial organisms. These families include Major facilitator superfamily (MFS), the small multidrug resistance family (SMR), resistance nodulation cell division family (RND), the ATP binding cassette family (ABC), The multidrug and toxic compound extrusion family (MATE) [66].
Reduced permeability: Several antibiotics used in the clinical settings are mostly intracellular bacterial target-based antibiotics. In case of gram-negative bacteria, the target is in the cytoplasmic membrane (the inner membrane). In order to attain an effective antimicrobial effect, the antibiotic compound must reach to its target site. Bacteria have developed such mechanisms that inhibit the antibiotics from reaching to its target site by decreasing their uptake [67]. These mechanisms are particularly important in the gram-negative bacteria that limit the influx of substances from external side. The outer membrane act as first line of defense inhibits the transport of toxic materials and antimicrobials inside the cell. The hydrophilic molecule such as β-Lactams, tetracycline and some fluroquinolones are particularly affected by changes in permeability of outer membrane as they often use water filled diffusion channels known as porins [67]. There are several types of porins classified on the basis of their structures (monomeric or trimeric). The best characterized porins are three major protein produced by coli known as (Omp F, Omp C, PhoE) and the Pseudomonas aeruginosa Opr D (also known as protein D2) are classical examples of porin mediated resistance [68].
Modification of antibiotic target: Natural alteration or acquired variations in the target sites of antimicrobial that prevent the binding of antibiotic to their target site is a common mechanism of resistance. The change in the target site results from spontaneous mutation of a bacterial gene on the chromosome. Since antibiotics have very specific target sites, a very minute changes in the target sites also disturb the working of antibiotics.
Alteration of 30S or 50S ribosomal subunit: The ribosome deal with the resistance of drugs that effect the protein synthesis i.e. macrolides, chloramphenicol, tetracycline and aminoglycosides. The aminoglycosides attach to the 30S subunit whereas macrolides, chloramphenicol, streptogramin B, and lincosamide bind with the 50S subunit to inhibit the protein synthesis [65,69].
Modification in Penicillin Binding Proteins (PBP): The variation in PBP is one of the best mechanisms of resistance in gram positive bacteria on the other hand the production of β-Lactamases is a mechanism of resistance in gram negative bacteria. The alteration in PBPs leads to decrease in the affinity of β-lactam antibiotics. The resistance to ampicillin in Enterococcus faecium and resistance to penicillin in pneumoniae is only due to this mechanism. Whereas the resistance in Staphylococcus aureus is due to the presence mobile genetic elements “Staphylococcal cassette chromosome mec” on the chromosome of Staphylococcus aureus that contain methicillin resistant gene mecA [63,70,71]. MECA gene is responsible for the synthesis of PBP2a protein, a novel penicillin binding protein that overcome the effect of Staphylococcal PBP. The Staphylococcus aureus strains that are resistant to penicillin also show resistance to all β-lactam antibiotics, tetracycline, streptomycin and in few cases to erythromycin [72].
Enzymatic degradation
There are three types of enzymes that prevent antibiotics to perform their functions such as β-Lactamases, Aminoglycoside modifying enzymes (AME) and chloramphenicol acetyltransferases (CAT) [73]. β-Lactamases inactivates almost all β-Lactam antibiotics that contain amide and ester bonds for example Penicillin, carbapenems, monobactam etc. Up till now around 300 β-Lactamases are identified. According to Ambler (Structural) classification system there are 4 classes of β-Lactamases such as Class A, Class B, Class C and Class D β-Lactamases [70]. The aminoglycoside modified enzymes AME are phosphoryl-transferase, nucleotidyl transferase or adenylyl transferase. These enzymes decrease the attraction of molecule and hence a result inhibit the attachment of molecule with 30S ribosomal subunit. The (AME) cause extended spectrum resistance to aminoglycoside and fluroquinolones. The resistance due to AME is mostly observed in the Staphylococcus aureus and S. pneumoniae strains [74]. The chloramphenicol acetyltransferase (CAT) enzyme is found in some gram positive, gram negative and in some strains of Hemophilus influenza strains. Due to presence of CAT enzyme the chloramphenicol is unable to bind with 50S ribosomal subunit [75].
Alternative to Antibiotics
The rapid rise of antibiotic resistance is very alarming condition for the scientists and health care professional who must need to develop the new antimicrobials for the treatment of drug resistant bacteria. Now a day’s scientists are continuously in search of various alternatives to the antibiotics that will help to combat the superbugs. Some of the alternatives are given below.
Antimicrobial photodynamic therapy
A rise in the antibiotic resistance in the pathogens also increase the morbidity and mortality as well as the cost of hospitals [76]. In the recent years the photodynamic therapy has been appeared as a non-invasive alternative treatment option for various infections caused by bacteria, fungi and viruses [77] The therapy is defined as oxygen dependent photochemical reaction that occurs upon light mediated activation of a photosynthesizing compounds leading to the generation of cytotoxic reactive oxygen species predominantly singlet oxygen [78].
There are three essential materials required for the PDT that are visible light, a photosensitizer and oxygen. An important thing is photosensitizer is harmless unless it is not exposed to the visible light [79]. When photosensitizer is exposed to a visible light of wavelength the photosensitizer will excite to a triplet energy state. The reaction that takes place between the triplet energy states photosensitizer and biomolecules leads to the cytotoxicity and antibacterial activity of PDT [80]. The triplet photosensitizer energy states initiate with one of possible two mechanisms known as type 1 and type 2 mechanism [81]. In type 1 mechanism, the transmission of electrons takes place between the excited photosensitizer and organic component of the cells. As a result of this interaction a highly reactive free radical species is produced that will react with oxygen molecule to from reactive oxygen species (ROS) such as superoxide, hydrogen peroxide and hydroxyl radical. The ROS attack on the cell membrane and cause irreversible damage to the cell membrane by the peroxidation of cell membrane components. IN type 2 mechanism, the excited photosensitizer reacts with oxygen to form a singlet oxygen that is a highly reactive form of oxygen. The singlet oxygen molecule causes oxidative damage to the cell membrane or cell wall [82,83].
Antimicrobial photodynamic therapy for infections has an edge that it eliminates the microorganisms independently of their antimicrobial resistance patterns and without any accurate microbial diagnosis. The APDT also has an advantage over other therapies that it has a very broad-spectrum activity, a very quick response time, very low chances of adverse side effects and small cost of treatment [84]. Several options are under study for the treatment of antibiotic resistant bacteria in near future. Among them one option is the combination of aPDT with the conventional antibiotics to gain the synergistic therapeutic effects or to control the antibiotic resistance [85,86]. Some of the examples of photosensitizers are given in the (Table 3).
Table 3: Photoinactivation of drug resistant pathogens.

Medicinal Plants
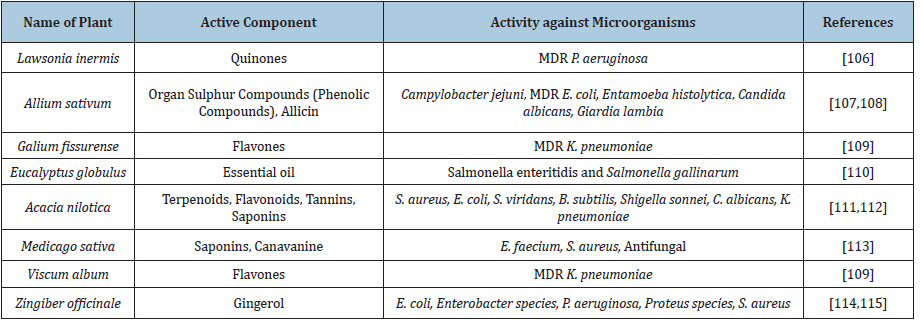
In the history of mankind plants played an exclusive role in providing food, drugs, shelter, clothing’s etc. to humans [87-94]. For the discovery of new drugs, the natural compounds explore broadly [95]. Indeed, plants have been use as a source of medicine for more than 5000 years ago [96]. Various studies showed that in the past humans have been used the natural compounds to fend off the infections [97]. The most powerful and promising components of plants are secondary metabolites on which the humans depend [98]. The secondary metabolites are heterogenous group of naturally occurring compounds that are used to treat various diseases [99]. Plants prepare the secondary metabolites (small organic molecules) that are not essential for their growth and development but require for the reproduction and defense against bacteria, fungus, virus, vertebrates etc. These secondary metabolites have strong potential to work as drug [100,101]. The secondary metabolites have been examined since 1850s. On the basis of chemical composition (Containing Nitrogen or not), chemical structure (have rings or contain a sugar) and their solubility the secondary metabolites are classified into 3 categories alkaloids, terpenes and phenolics [102]. The major part of compounds obtained from plants are phytochemicals and secondary metabolites that play an important role as antimicrobials and antivirals and categorized into several groups such as alkaloids, phenolics, polyphenols, flavonoids, quinones, tannins, coumarins, terpenes, lectins, polypeptides and saponins etc. [103-105]. Various studies showed that medicinal plants have antimicrobial activities. Some of the studies are shown in the (Table 4).
Table 4: Medicinal plants showing antibacterial activity against MDR pathogens.
Probiotics as alternatives
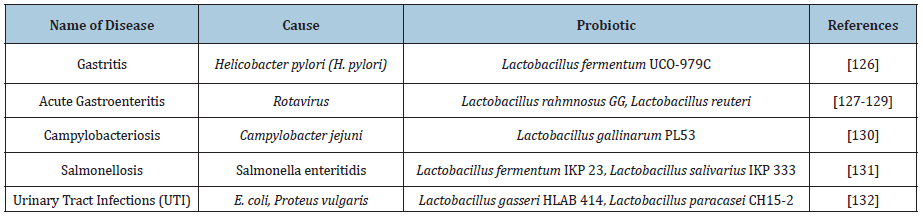
During recent years probiotics get the attention of both consumers and research. Increasing clinical evidences revealed that the use of probiotics induces health benefits particularly in diarrheal diseases. Probiotics are defined as “The live microbes when given in an adequate amount provides health benefits to the host” [106-116]. Probiotics are regulated as dietary supplements and foods consists of yeast or bacteria. They are also available in the form of capsule, tablets, powder forms and also in dairy products such as yogurt etc. the most commonly used probiotics are lactic acid bacteria such as Lactobacillus and Bifidobacterium species. The yeast Saccharomyces bouvardia also exhibit the probiotic potential. It is important to note that the probiotic effects are specific to one strain, so the health effect applicable to one strain is not attributed to the other strains even within one species. Therefore, the generalization about the potential health benefits should not be made [117-120]. The mechanism of action of probiotics is not clearly known yet. However, several mechanisms have been proposed for them. As described previously the probiotics used most frequently are lactic acid bacteria and Bifidobacterium species. These bacteria produce lactic acid, acetic acid and propionic acid that results in the decline in the intestinal pH and inhibit the growth of several pathogenic bacteria [119,121].
The probiotic strain with this ability is Lactobacillus species strain GG that secrete a low molecular weight compound which has broad spectrum activity that inhibits gram positive, gram negative and anaerobic bacteria [122]. Probiotics also reduce the colonization of various pathogenic microbes in urinary and intestinal tract by adhesion to the epithelial cells [123]. Some studies showed that Lactobacillus has ability to inhibit the attachment of E. coli, P. aeruginosa and K. pneumoniae with the uroepithelial and intestinal epithelial cells [124,125]. Probiotics are used for the treatment and prevention of various medical conditions and also to promote the general health. Several studies described that probiotics can also use as alternatives to antibiotics as shown in the (Table 5).
Table 5: Probiotic potential of various Lactobacillus strains against drug resistant pathogens.
Nanoparticles as antimicrobial
In the present time the development in the field of nanoscience and nanotechnology has developed the interest of scientists in the synthesis of nanosized inorganic and organic particles that have a wide range of applications in industrial, medical and therapeutics, synthetic textile, and food packaging products [126-133]. Nanoparticles normally fall in the range of 1-100 nm in diameter. With the decrease in their size to atomic level the properties of nanoparticles also changed. The nanoparticles have unique physico-chemical, optical and chemical properties that can be used according to the required applications [134]. Furthermore, the biological process also takes place at nanoscale and due to their amenability to the biological functionalization the nanoparticles have an important application in the field of medicines [135].
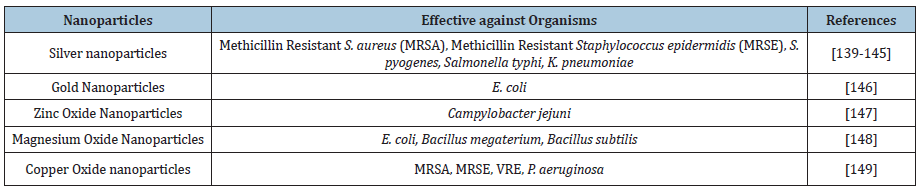
Now a days the metallic nanoparticles are systematically examined and broadly investigated as potential source of antimicrobials. The antimicrobial activity of nanoparticles is known to be a function of the surface area in contact with the microorganisms. The small size and high surface to volume ratio increase the interaction of nanoparticles to attach with the microorganisms and also to carry a large amount of antibiotics. The metal nanoparticles in combination with antibiotics show an immense application in water treatment, synthetic textile, biomedical and surgical devices, food processing and packaging [136]. There are various mechanisms that describe the inhibitory effect of silver nanoparticles on bacteria. The affinity of silver towards the Sulfur and phosphorous is a key element of the antimicrobial effect. Due to profusion of sulfur proteins in the cell membrane, the silver nanoparticles interact with the sulfur containing amino acids inside or outside the cell membrane that disturbs the bacterial cell viability. The nanoparticles can also interact with the phosphorous moieties in the DNA, resulting in inactivation of DNA replication, leading to inhibition of enzyme functions [137,138]. Different studies described that several nanoparticles exhibit effective antimicrobial activity against pathogenic organisms. Some of them are shown in the Table 6 below:
Table 6: Nanoparticles exhibit antibacterial activity against drug resistant pathogens.
Conclusion
To conclude our review, it reveals that antibiotic resistance is on rise in all over the world. The consequences of antibiotic resistant bacterial infections are prolonged stay at hospitals, treatment failures and high economic burden [139-149]. The public health experts need to develop such a surveillance system coordinated at the national and international levels, ongoing analysis and a mandatory reporting system for antibiotic resistance. Both domestic and global policies need to be conventional and adhere to stop the overuse and misuse of antibiotics.
References
- Levy SB, B Marshall (2004) Antibacterial resistance worldwide: Causes, challenges and responses. Nat Med 10(12): S122-S129.
- Levy SB (2013) The antibiotic paradox: How miracle drugs are destroying the miracle. JAMA 270(3): 384-385.
- Aminov RI (2010) A brief history of the antibiotic era: Lessons learned and challenge for the future. Front Microbiol 1: 134.
- Davies J, D Davies (2010) Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev 74(3): 417-433.
- WHO (2014) Antimicrobial resistance: Global report on surveillance? Switzerland, pp. 1-232.
- Editorial (2013) The antibiotic alarm. Nat 495(7440): 141.
- WHO (2015) Global action plan on antimicrobial resistance? Switzerland.
- Gandra S, Barter D, Laxminarayan R (2014) Economic burden of antibiotic resistance: how much do we really know? Clin Microbiol Infect 20(10): 973-980.
- Falagas ME, Giannoula ST, Drosos EK, Konstantinos ZV (2014) Deaths attributable to carbapenem-resistant Enterobacteriaceae Emerging Infectious Diseases 20(7): 1170-1175.
- Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, et al. (2012) Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clinic Microbiol Infect 18(3): 268-281.
- Rothman L (2013) Antibiotic resistance threats in the united states. USA, pp. 1-118.
- Akova M (2016) Epidemiology of antimicrobial resistance in bloodstream infections. Virulence 7(3): 252-266.
- Mühlen S, Dersch P (2015) Anti-virulence strategies to target bacterial infections, in How to overcome the antibiotic crisis. Curr Top Microbiol Immunol 398: 147-183.
- Sengupta S, Chattopadhyay MK, Grossart HP (2013) The multifaceted roles of antibiotics and antibiotic resistance in nature. Front Microbiol 4: p.47.
- Spellberg B, Gilbert DN (2014) The future of antibiotics and resistance: A tribute to a career of leadership by John Bartlett. Clin Infect Dis 59(suppl_2): S71-S75.
- Hoge CW, Gambel JM, Srijan A, Pitarangsi C, Echeverria P, et al. (1998) Trends in antibiotic resistance among diarrheal pathogens isolated in Thailand over 15 years. Clin Infect Dis 26(2): 341-345.
- Gootz TD (1990) Discovery and development of new antimicrobial agents. Clin Microbiol Rev 3(1): 13-31.
- Jevons MP (1961) Celbenin resistant staphylococci. Br Med J 1(5219): 124.
- Lowy FD (2003) Antimicrobial resistance: The example of Staphylococcus aureus. J Clin Invest 111(9): 1265-1273.
- Appelbaum PC (2006) The emergence of vancomycin‐intermediate and vancomycin‐resistant Staphylococcus aureus. Clin Microbiol Infect 12: 16-23.
- Hansman D, Bullen MM (1967) A resistant pneumococcus. Lancet 290(7509): 264-265.
- Ashford W, Golash R, Hemming V (1976) Penicilunase-producing neisseria gonorrhή. The Lancet 308(7987): 657-658.
- Leclercq R, Derlot E, Duval J, Courvalin P (1988) Plasmid-mediated resistance to vancomycin and teicoplanin in Enterococcus faecium. N Engl J Med 319(3): 157-161.
- Knothe H, Shah P, Krcmery V, Antal M, Mitsuhashi S, et al. (1983) Transferable resistance to cefotaxime, cefoxitin, cefamandole and cefuroxime in clinical isolates of Klebsiella pneumoniae and Serratia marcescens. Infect 11(6): 315-317.
- Yigit H, Queenan AM, Anderson GJ, Sanchez AD, Biddle JW, et al. (2001) Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother 45(4): 1151-1161.
- CDC (2002) Staphylococcus aureus resistant to vancomycin-United States, 2002. MMWR 51(26): 565-567.
- Mangili A, Bica I, Snydman DR, Hamer DH (2005) Daptomycin-resistant, methicillin-resistant Staphylococcus aureus Clinic Infect Diseas 40(7): 1058-1060.
- CDC (2007) Update to CDC's sexually transmitted diseases treatment guidelines, 2006: fluoroquinolones no longer recommended for treatment of gonococcal infections. MMWR 56(14): 332-336.
- Olusegun OS, Doug H, Sean S, Kim T, Kathie AR, et al. (2012) Emergence of increased azithromycin resistance during unsuccessful treatment of Neisseria gonorrhoeae infection with azithromycin (Portland, OR, 2011). Sex Transm Dis 39(11): 877-879.
- Humphries RM, Shangxin Y, Peera H, Kevin W W, Janet AH, et al. (2015) First report of ceftazidime-avibactam resistance in a KPC-3-expressing Klebsiella pneumoniae isolate. Antimicrob Agents Chemother 59(10): 6605-6607.
- McEwen SA, Cray PJF (2002) Antimicrobial use and resistance in animals. Clin Infect Dis 34(3): S93-S106.
- Witte W (1998) Medical consequences of antibiotic use in agriculture. American Association for the Advancement of Science 279(5353): 996-997.
- Garau J, Mariona X, Mónica RC, Josep RGV, Ignacio C, et al. (1999) Emergence and dissemination of quinolone-resistant escherichia coli in the community. Antimicrob Agents Chemother 43(11): 2736-2741.
- Read AF, Woods RJ (2014) Antibiotic resistance management. Evol Med Public Health 2014(1): 147.
- Michael CA, Dominey H, Labbate M (2014) The antimicrobial resistance crisis: causes, consequences, and management. Front Public Health 2: 145.
- Mor A, Trine F, Reimar WT, Alessandro O, Peter R, et al., (2015) Antibiotic use varies substantially among adults: a cross-national study from five European Countries in the ARITMO project. Infection 43(4): 453-472.
- Ternhag A, J Hellman (2013) More on US outpatient antibiotic prescribing, 2010. N Engl J Med 369(12): 1175-1176.
- LIM (2014) L'Agence nationale de sécurité du médicament et des produits de santé. SOINS. CADRES (89): S27-S28.
- Baclet N, Grégoire F, Serge A, Laurie F, Eric S, et al. (2017) Explicit definitions of potentially inappropriate prescriptions of antibiotics in older patients: A compilation derived from a systematic review. International Journal of Antimicrobial Agents 50(5): 640-648.
- Struelens MJ (1998) The epidemiology of antimicrobial resistance in hospital acquired infections: Problems and possible solutions. BMJ 317(7159): 652-654.
- Muhammad A, Muhammad A, Anum S, Shane S (2017) Investigation of antimicrobial use at a tertiary care hospital in southern Punjab, Pakistan using WHO methodology. Antimicrob Resist Infect Control 6(1): 41.
- Morgan DJ, Iruka NO, Ramanan L, Eli NP, Scott W, et al. (2011) Non-prescription antimicrobial use worldwide: A systematic review. The Lancet infectious diseases 11(9): 692-701.
- Hussain T (2015) Pakistan at the verge of potential epidemics by multi-drug resistant pathogenic bacteria. Adv Life Sci 2(2): 46-47.
- Ashraf F, Faisal I, Farah A, Hassan I (2017) Antibiotic dispensing and prescription pattern in pharmacies of Islamabad and Rawalpindi: Pakistan. International Journal of Collaborative Research on Internal Medicine & Public Health 9(5).
- Klein EY, Thomas PVB, Elena MM, Suraj P, Sumanth G, et al. (2018) Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proceedings of the National Academy of Sciences 115(15): E3463-E3470.
- Lushniak BD (2014) Antibiotic resistance: A public health crisis. Public Health Reports 129(4): 314-316.
- Viswanathan V (2014) Off-label abuse of antibiotics by bacteria. Gut Microbes 5(1):3-4.
- Thamlikitkul V (1988) Antibiotic dispensing by drug store personnel in Bangkok, Thailand. J Antimicrob Chemoth 21(1): 125-131.
- Okeke IN, Lamikanra A, Edelman R (1999) Socioeconomic and behavioral factors leading to acquired bacterial resistance to antibiotics in developing countries. Emerg Infect Dis 5(1): 18-27.
- Lansang MA, Aquino RL, Tupasi TE, Mina VS, Salazar LS, et al. (1990) Purchase of antibiotics without prescription in Manila, the Philippines. Inappropriate choices and doses. J Clin Epidemiol 43(1): 61-67.
- Bank W (2016) Physicians (per 1,000 people). USA.
- https://www.cdc.gov/
- Bartlett JG, Gilbert DN, Spellberg B (2013) Seven ways to preserve the miracle of antibiotics. Clinical Infectious Diseases 56(10): 1445-1450.
- Gross M (2013) Antibiotics in crisis. Curr Biol 23(24): R1063-1065.
- Golkar Z, Bagasra O, Pace DG (2014) Bacteriophage therapy: A potential solution for the antibiotic resistance crisis. J Infect Dev Ctries 8(2): 129-136.
- Rossolini GM, Fabio A, Patrizia P, Simona P (2014) Update on the antibiotic resistance crisis. Curr Opin Pharmacol 18: 56-60.
- WHO (2018) Global antimicrobial resistance surveillance system (GLASS) report: Early implementation 2017-2018? pp. 1-268.
- Maragakis LL, Perencevich EN, Cosgrove SE (2008) Clinical and economic burden of antimicrobial resistance. Expert Rev Anti-Infect Ther 6(5): 751-763.
- Brad S, Martin B, Robert JG, Helen WB, John SB, et al. (2011) Combating antimicrobial resistance: Policy recommendations to save lives. Clinic Infect Dis 52(5): S397-S428.
- Ventola CL (2015) The antibiotic resistance crisis: part 1: Causes and threats. P T 40(4): 277-283.
- Munita JM, CA Arias (2016) Mechanisms of antibiotic resistance. Virulence Mechanisms of Bacterial Pathogens pp. 481-511.
- Manson JM, Hancock LE, Gilmore MS (2010) Mechanism of chromosomal transfer of Enterococcus faecalis pathogenicity island, capsule, antimicrobial resistance, and other traits. Proc Natl Acad Sci U S A 107(27): 12269-12274.
- Džidić S, Šušković, Kos B (2008) Antibiotic resistance mechanisms in bacteria: biochemical and genetic aspects. Food Technology & Biotechnology 46(1).
- Wise R (1999) A review of the mechanisms of action and resistance of antimicrobial agents. Can Respir J 6: 20A-2A.
- Lambert PA (2002) Mechanisms of antibiotic resistance in Pseudomonas aeruginosa. J R Soc Med 95(Suppl 41): 22-26.
- Piddock LJ (2006) Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clinic Microbiol Rev 19(2): 382-402.
- Pagès JM, James CE, Winterhalter M (2008) The porin and the permeating antibiotic: A selective diffusion barrier in Gram-negative bacteria. Nat Rev Microbiol 6(12): 893-903.
- Nikaido H, Vaara M (2003) Molecular basis of bacterial outer membrane permeability revisited. Microbiol Rev 67(4): 593-656.
- Tenover FC (2006) Mechanisms of antimicrobial resistance in bacteria. Am J Med 119(6): S3-S10.
- Alekshun MN, SB Levy (2007) Molecular mechanisms of antibacterial multidrug resistance. Cell 128(6): 1037-1050.
- Hiramatsu K (2001) The emergence and evolution of methicillin-resistant Staphylococcus aureus. Trends Microbiol 9(10): 486-493.
- Grundmann H (2006) Emergence and resurgence of meticillin-resistant Staphylococcus aureus as a public-health threat. Lancet 368(9538): 874-885.
- Goering R (2012) Attacking the enemy: antimicrobial agents and chemotherapy: macrolides. Medical Microbiology 5: 447-489.
- Strateva T, Yordanov (2009) Pseudomonas aeruginosa a phenomenon of bacterial resistance. J Med Microbiol 58(9): 1133-1148.
- Tolmasky ME (2000) Bacterial resistance to aminoglycosides and beta-lactams: the Tn1331 transposon paradigm. Front Biosci 5(10): 11.
- Geralde MC (2017) Pneumonia treatment by photodynamic therapy with extracorporeal illumination‐an experimental model. Physiol Rep 5(5): e13190.
- Kornman KS, Page RC, Tonetti MS (1997) The host response to the microbial challenge in periodontitis: assembling the players. Periodontol 14(1): 33-53.
- Ochsner M (1997) Photophysical and photobiological processes in the photodynamic therapy of tumours. J Photochem Photobiol B 39(1): 1-18.
- Fekrazad R, Hadi, Sara, Parisa (2014) The effect of antimicrobial photodynamic therapy with radachlorin® on Staphylococcus aureus and Escherichia coli: an in vitro J Lasers Med Sci 5(2): 82.
- Maisch T (2007) Anti-microbial photodynamic therapy: useful in the future? Lasers Med Sci 22(2): 83-91.
- Gursoy H, Ceyda, Tomruk (2013) Photodynamic therapy in dentistry: a literature review. Clinical Oral Investigations 17(4): 1113-1125.
- Foote CS (1991) Definition of type I and type II photosensitized oxidation. Photochemistry and Photobiology 54(5): 659-659.
- Sharman WM, Allen CM, Lier JE (1999) Photodynamic therapeutics: basic principles and clinical applications. Drug Discovery Today 4(11): 507-517.
- Riordan K, Akilov OE, Hasan T (2005) The potential for photodynamic therapy in the treatment of localized infections. Photodiagnosis and Photodynamic Therapy 2(4): 247-262.
- Pérez LV, Luna, Vernoica, Antonia, Yoland, et al. (2017) Bactericidal effect of photodynamic therapy, alone or in combination with mupirocin or linezolid, on Staphylococcus aureus. Frontiers in Microbiology 8: 1002.
- Wozniak A, Grinholc M (2018) Combined antimicrobial activity of photodynamic inactivation and antimicrobials-state of the art. Frontiers in Microbiology 9: 930.
- Li J, Liu, Xiayogoan, Yanhoji, Qin, et al. (2020) Antimicrobial photodynamic therapy against multidrug-resistant Acinetobacter baumannii clinical isolates mediated by aloe-emodin. An In vitro Study 29: 101632.
- Liu C (2016) Photodynamic inactivation of antibiotic-resistant bacteria and biofilms by hematoporphyrin monomethyl ether. Lasers in Medical Science 31(2): 297-304.
- Paschoal MA, Meng, Pinto (2015) Photodynamic antimicrobial chemotherapy on Streptococcus mutans using curcumin and toluidine blue activated by a novel LED device. Lasers in Medical Science 30(2): 885-890.
- Wilder S (2002) Photoeradication of Helicobacter pylori using 5‐aminolevulinic acid: Preliminary human studies. Lasers in Surgery and Medicine. Lasers Surg Med 31(1): 18-22.
- Ganz RA (2005) Helicobacter pylori in patients can be killed by visible light. Lasers Surg Med 36(4): 260-265.
- Lembo AJ (2009) Treatment of Helicobacter pylori infection with intra‐gastric violet light phototherapy: a pilot clinical trial. Lasers Surg Med 41(5): 337-344.
- Mai B, Gao, Min, Wang, Zhang, et al. (2017) Photodynamic antimicrobial chemotherapy for Staphylococcus aureus and multidrug-resistant bacterial burn infection in vitro and in vivo. Int J Nanomedicine 12: 5915.
- Sueoka K, Chikama, Latief, Jiaeka, Obana, et al. (2018) Time-dependent antimicrobial effect of photodynamic therapy with TONS 504 on Pseudomonas aeruginosa. Lasers in Medical Science 33(7): 1455-1460.
- Chandra H. Archana, Parul, Babita, Anant, et al. (2017) Antimicrobial resistance and the alternative resources with special emphasis on plant-based antimicrobials-a review. Plants 6(2): 16.
- Robinson MM, Zhang (2011) The world medicines situation 2011, traditional medicines: Global situation, issues and challenges. World Health Organization, Geneva, Switzerland, pp. 1-2.
- Newman DJ, Cragg G (2016) Natural products as sources of new drugs from 1981 to 2014. J Nat Prod 9(3): 629-661.
- Sen S, Chakraborty R (2011) Challenges and opportunities in the advancement of herbal medicine: India’s position and role in a global context. Journal of Herbal Medicine 1(3-4): 67-75.
- Moloney MG (2016) Natural products as a source for novel antibiotics. Trends Pharmacol Sci 37(8): 689-701.
- Boy HIA, Rutilla, Santos, Alicia, Tooba, et al. (2018) Recommended medicinal plants as source of natural products: a review. Digital Chinese Medicine 1(2): 131-142.
- Mawalagedera SM, Damien LC, Anne CG, Nina R, Matthew RE (2019) Combining evolutionary inference and metabolomics to identify plants with medicinal potential. Frontiers in Ecology and Evolution 7: 267.
- Kabera JN (2014) Plant secondary metabolites: biosynthesis, classification, function, and pharmacological properties. J Pharm Pharmacol 2: 377-392.
- Alamgir A (2017) Therapeutic use of medicinal plants and their extracts. Springer, Germany, Volume 1, pp. 177-293.
- Connor SE (2015) Engineering of secondary metabolism. Annual Review of Genetics 49: 71-94.
- Katz L, Baltz RH (2016) Natural product discovery: past, present, and future. J Ind Microbiol Biotechnol 43(2-3): 155-176.
- Habbal O, Hasson SS, Hag, Mahrooqi, Bimani, et al. (2011) Antibacterial activity of Lawsonia inermis Linn (Henna) against Pseudomonas aeruginosa. Asian Pacific Journal of Tropical Biomedicine 1(3): 173-176.
- Lu X, Jamie MF, Aston E, Lin M, Michael EK, et al. (2011) Investigating antibacterial effects of garlic (Allium sativum) concentrate and garlic-derived organosulfur compounds on Campylobacter jejuni by using Fourier transform infrared spectroscopy, raman spectroscopy, and electron microscopy. Applied and Environmental Microbiology 77(15): 5257-5269.
- Ankri S, Mirelman (1999) Antimicrobial properties of allicin from garlic. Microbes Infects 1(2): 125-129.
- Özçelik B (2008) Antimicrobial activity of flavonoids against extended-spectrum β-lactamase (ESβL)-producing Klebsiella pneumoniae. Tropical Journal of Pharmaceutical Research 7(4): 1151-1157.
- Yasmin S, Nawaz M, Aftab AA, Kamran A, Ullah N, et al. (2019) Antibiotic susceptibility pattern of Salmonellae isolated from poultry from different districts of Punjab, Pakistan. Pak Vet J 40: 98-102.
- Banso A (2009) Phytochemical and antibacterial investigation of bark extracts of ACACIA nilotica. Journal of Medicinal Plants Research 3(2): 082-085.
- Riaz S, Faisal M, Hasnain S, Naveed AK (2011) Antibacterial and cytotoxic activities of Acacia nilotica Lam (Mimosaceae) methanol extracts against extended spectrum Beta-Lactamase producing Escherichia coli and Klebsiella Tropical Journal of Pharmaceutical Research 10(6): 785-791.
- Aliahmadi A, Rasoul, Giti, Fatemeh, Alireza (2012) Identification and primary characterization of a plant antimicrobial peptide with remarkable inhibitory effects against antibiotic resistant bacteria. African Journal of Biotechnology 11(40): 9672-9676.
- Adeshina G, Jibo S, Agu VE, Joseph (2011) Antibacterial activity of fresh juices of Allium cepa and Zingiber officinale against multidrug resistant bacteria. International Journal of Pharma and Bio Sciences 2(2): 289-295.
- Karuppiah P, Rajaram S (2012) Antibacterial effect of Allium sativum cloves and Zingiber officinale rhizomes against multiple-drug resistant clinical pathogens. Asian Pac J Trop Biomed 2(8): 597-601.
- Joint F (2002) The evaluation of probiotics in food. London, Food and Agriculture Organization of the United Nations, WHO, Canada, pp. 1-12.
- Senok AA, Ismaeel, Botta (2005) Probiotics: facts and myths. Clin Microbiol Infect 11(12): 958-966.
- Santosa SE, Farnworth, Jones PJ (2006) Probiotics and their potential health claims. Nutr Rev 64(6): 265-274.
- Alvarez OM, Oberhelman (2001) Probiotic agents and infectious diseases: a modern perspective on a traditional therapy. Clin Infect Dis 32(11): 1567-1576.
- Pham M, Lemberg DA, Day AS (2008) Probiotics: sorting the evidence from the myths. Medical Journal of Australia 188(5): 304-308.
- Doron S, Gorbach SL (2006) Probiotics: their role in the treatment and prevention of disease. Expert Rev Anti Infect Ther 4(2): 261-275.
- Silva M, Jacobus NV, Deneke, Gorbach (1987) Antimicrobial substance from a human Lactobacillus Asian Pac J Trop Biomed 31(8): 1231-1233.
- Macintyre A, Childscymet (2005) Probiotics The benefits of bacterial cultures. Comprehensive Therapy 31(3): 181-185.
- Chan RC, Reid G, Irvin RT, Bruce AW, Costerton JW (1985) Competitive exclusion of uropathogens from human uroepithelial cells by Lactobacillus whole cells and cell wall fragments. Infects Immun 47(1): 84-89.
- Mack DR, Michail, Wei, Dougall, Hollingsworth (1999) Probiotics inhibit enteropathogenic coli adherence in vitro by inducing intestinal mucin gene expression. Am J Physiol 276(4): G941-G950.
- Garcia CV, Villena, Ilabbaco, Zelaya, Monje, et al. (2018) Lactobacillus fermentum UCO-979C beneficially modulates the innate immune response triggered by Helicobacter pylori infection in vitro. Benef Microbes 9(5): 829-841.
- Kaila M, Laine, Soppi, Isilouri (1992) Enhancement of the circulating antibody secreting cell response in human diarrhea by a human Lactobacillus Clinical Trial 32(2): 141-144.
- Saavedra JM (1994) Feeding of Bifidobacterium bifidum and Streptococcus thermophilus to infants in hospital for prevention of diarrhoea and shedding of rotavirus. The lancet 344(8929): 1046-1049.
- Sugita T (1994) Efficacy of Lactobacillus preparation biolactis powder in children with rotavirus Jpn J Pediatr 47: 2755-2762.
- Khan M (2019) Effect of newly characterized probiotic lactobacilli on weight gain, immunomodulation, and gut microbiota of campylobacter jejuni challenged broiler chicken. Pakistan Veterinary Journal 39(4).
- Khan I (2019) Isolation and in vitro characterization of anti-Salmonella enteritidis probiotic potential of indigenous lactobacilli from poultry. Pak Vet J 39(4): 563-567.
- Shim YH, Lee (2016) Antimicrobial activity of lactobacillus strains against uropathogens. Pediatr Int 58(10): 1009-1013.
- Gajjar P (2009) Antimicrobial activities of commercial nanoparticles against an environmental soil microbe, Pseudomonas putida J Biol Eng 3(1): 1-13.
- Feynman R (1991) There's plenty of room at the bottom. Science 254: 1300-1301.
- Whitesides GM (2003) The right size in nanobiotechnology. Nat Biotechnol 21(10): 1161-1165.
- Martinez GF, Peggy, Facundo, Hoaracio, Yossef, et al. (2010) Synthesis, characterization, and evaluation of antimicrobial and cytotoxic effect of silver and titanium nanoparticles. Nanomedicine: Nanotechnology, Biology and Medicine 6(5): 681-688.
- Gupta A, Silver (1998) Silver as a biocide: will resistance become a problem? Nat Biotechnol 16(10): 888-888.
- Matsumura Y, Kuniaki Y, Shinichi K, Tetsuaki T (2003) Mode of bactericidal action of silver zeolite and its comparison with that of silver nitrate. Appl Environ Microbiol 69(7): 4278-4281.
- Yamanaka M, Hara, Kudo (2005) Bactericidal actions of a silver ion solution on Escherichia coli, studied by energy-filtering transmission electron microscopy and proteomic analysis. Appl Environ Microbiology 71(11): 7589-7593.
- Lara HH, Nilda, Carmen, Cristina (2010) Bactericidal effect of silver nanoparticles against multidrug-resistant bacteria. World Journal of Microbiology and Biotechnology 26(4): 615-621.
- Shahverdi AR, Ali (2007) Synthesis and effect of silver nanoparticles on the antibacterial activity of different antibiotics against Staphylococcus aureus and Escherichia coli. Nanomedicine: Nanotechnology, Biology and Medicine 3(2): 168-171.
- Yoon K, Hoon, Park, Hwang (2007) Susceptibility constants of Escherichia coli and Bacillus subtilis to silver and copper nanoparticles. Sci Total Environ 373(2-3): 572-575.
- Sarkar S, Jana (2007) Facile synthesis of silver nano particles with highly efficient anti-microbial property. Polyhedron 26(15): 4419-4426.
- Shrivastava S, Bera, Arnab, Singh G, Ramachandra R, et al. (2007) Characterization of enhanced antibacterial effects of novel silver nanoparticles. Nanotechnology 18(22): 225103.
- Nanda A, Saravanan (2009) Biosynthesis of silver nanoparticles from Staphylococcus aureus and its antimicrobial activity against MRSA and MRSE. Nanomedicine 5(4): 452-456.
- Cui Y, Zhao, Yue, Zhang, Jiang (2012) The molecular mechanism of action of bactericidal gold nanoparticles on Escherichia coli. Biomaterials 33(7): 2327-2333.
- Xie Y, He, Peter, Tony, Shi (2011) Antibacterial activity and mechanism of action of zinc oxide nanoparticles against Campylobacter jejuni. Applied and Environmental Microbiology 77(7): 2325-2331.
- Koper OB (2002) Nanoscale powders and formulations with biocidal activity toward spores and vegetative cells of bacillus species, viruses, and toxins. Current Microbiology 44(1): 49-55.
- Agarwala MB, Choudhury, Yadav (2014) Comparative study of antibiofilm activity of copper oxide and iron oxide nanoparticles against multidrug resistant biofilm forming uropathogens. Indian J Microbiol 54(3): 365-368.
© 2020 Muhammad Nawaz. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






