- Submissions

Full Text
Cohesive Journal of Microbiology & Infectious Disease
COVID-19 Pandemic Simulation Studies on the Transmissibility
Hyunjo Kim1* and Hoon Song J2
1School of pharmacy, South Korea
2Chairman, Asian Pacific Research Foundation for Infectious, ANSOP, South Korea
*Corresponding author: Hyunjo Kim, School of pharmacy, South Korea
Submission: May 02, 2020; Published: May 28, 2020

ISSN 2578-0190 Volume3 issues5
Abstract
COVID-19 was identified as the causative virus of pneumonia based on unknown etiology. COVID-19 has multiple characteristics distinct from other infectious diseases, including high infectivity during incubation, time delay between real dynamics and daily numbers of confirmed cases, and the intervention effects of quarantine and control measures. Public health concerns are being paid globally on how many people are infected and suspected reach to pandemics. Therefore, it is urgent to develop a mathematical model to estimate the transmissibility and dynamic of the transmission of the virus.
Keywords: COVID-19; Pandemic; Radiology; Machine learning; Mathematical model; Transmissibility
Introduction
Emerging infectious diseases (EIDs) and their determinants have recently attracted substantial scientific and popular attention. Over 75% of EIDs consist of zoonotic, Coronaviruses are capable of interspecies transmission [1-4]. Some of them have caused worldwide panic as emerging human pathogens in recent years, e.g., severe acute respiratory syndrome coronavirus (SARS-CoV) [5-10] and Middle East respiratory syndrome coronavirus (MERS-CoV) [11-18]. Coronavirus are a group of enveloped positive-stranded RNA viruses responsible for a variety of diseases in birds and mammals and belongs to the Coronavirus genus of the family [19-21]. Corona-viridae; one of its variants, named SARS virus, can cause severe acute respiratory syndrome (SARS). The Coronaviruses genome, ranging from 26 to 32 kilo-bases in length, is probably the largest viral RNA known. Previously, there are six Coronavirus known to cause human diseases and can be divided into low pathogenic and highly pathogenic Coronavirus. The low pathogenic Coronavirus, including 229E, HKU1, OC43 and NL63, account for 10% to 30% of upper respiratory tract infections and typically cause mild respiratory diseases [20-22]. In contrast, the highly pathogenic Coronavirus, including Severe Acute Respiratory Syndrome (SARS) and Middle East Respiratory Syndrome (MERS) Coronavirus, predominantly infect lower airways and cause fatal pneumonia [5,21]. These two highly pathogenic Coronavirus have posed a substantial threat to public health. World health organization (WHO) determined that the pathogen is novel coronavirus with many similarities to SARS virus. Subsequently, the full genomic sequence from Shanghai Public Health Clinical Center argued for a bat origin for the COVID-19 [23-33]. Despite several decades of research, specific vaccine or treatment for human Coronavirus is lack. In this review, we summarize the advance of the nature of the COVID- 19 and its clinical characteristics and therapeutics, which may be critical for the response to the COVID-19 outbreak. In order to assess their threat to humans, we explored to infer the potential hosts of coronaviruses using a dual-model approach with discriminant model achieved high accuracies in leave-one-out cross-validation of training data consisting of standard representative coronaviruses [34-37]. Predictions on chosen additional coronaviruses precisely conformed to conclusions or speculations by other researchers. The novel coronavirus disease 19 (COVID-19) is rapidly spreading with a rising death toll and transmission rate reported in high income countries rather than in low income countries. The overburdened healthcare systems and poor disease surveillance systems in resource-limited settings may struggle to cope with this COVID-19 outbreak and this calls for a tailored strategic response for these settings. Here, we recommend blockchain and artificial intelligence-coupled tracking systems for COVID-19 and other emerging infectious diseases. Prompt deployment and appropriate implementation of the proposed system have the potential to curb the transmissions of COVID-19 and the related mortalities, particularly in settings with poor access to laboratory infrastructure.
Materials and Methods
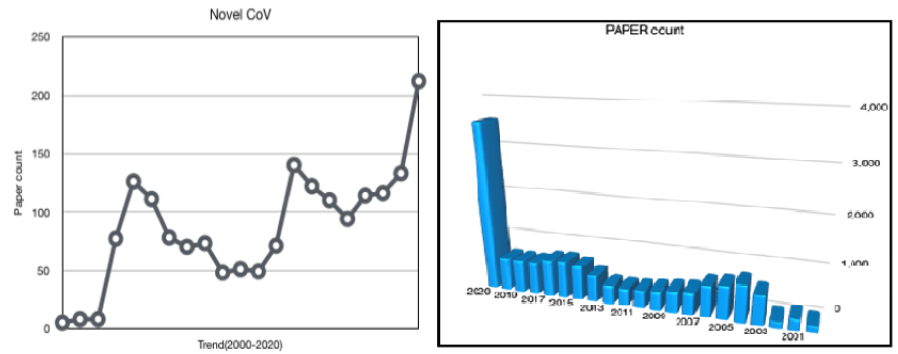
A literature search was performed using the PubMed database and the Cochrane library. Search terms included “novel coronavirus” and “2019-nCoV”. The MESH terms were: “novel” [All Fields] AND (“coronavirus”[MeSH Terms] OR (“2019-nCoV”[All Fields]) OR (“COVID-19” [All Fields]. The defined search period from November 30 2019 to March 18 2020 was selected to compare studies regarding first outbreaks and findings. Given the nature of the review, no ethics approval was required. The search was performed by two investigators. A total of 4,587 studies were identified (PubMed: 4,585, Cochrane: 2) in accordance with the results shown in Figure 1. Two investigators then reviewed these articles, initially by title and abstract and then in detail, using a customized data abstraction form. Studies were excluded if they had an incorrect subject matter, were duplications or review.
Figure 1: COVID-19 Research trend according to Meta-analysis.
Data source
Figure 2: Modified SEI/AR Scheme in this study.
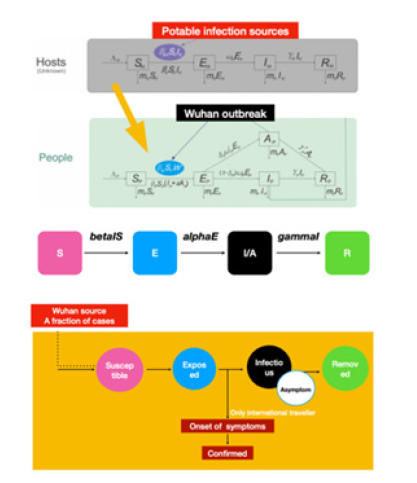
The reported cases of COVID-19, were collected for the modeling study from a published literature and Wikipedia coronavirus outbreak in South Korea. The epidemic curve from 20 January, 2020 to 18 April, 2020 was collected for our study, and the simulation time step was 1 day. We introduced the modified and simplified reservoir-people transmission network model from Chen (Figure 2) applying the assumption that COVID-19 might be imported to the South Korea in a short time. Therefore, we added the further assumptions as follows: During the outbreak period, the natural birth rate and death rate in the population was in a relative low level. Since there was no data on the proportion of asymptomatic infection of the virus, Since there was no evidence about the transmissibility of asymptomatic infection, we assumed that it could be died in the environment in a short time, but it could be stay for a longer time (14 days) in the unknown hosts.
Results
Characteristics of the novel Coronavirus 2019
Public health responses: A few cases of unidentified pneumonia with a history of exposure were reported at Wuhan, Hubei Province of China in December 2019. As is known SARS-CoV-2, a novel coronavirus was identified to be accountable for this SARS outbreak. Human to human transmission was confirmed in this disease named COVID-19 by WHO spread rapidly around. This COVID-19 resulted in a much lower-case fatality rate approximately 2.67% among the confirmed cases, compared with Severe Acute Respiratory Syndrome (SARS) and Middle East Respiratory Syndrome (MERS) [38-44]. According to the released official reports, the top four are fever, cough, short of breath, and chest tightness/pain. The major co-morbidities of the fatality cases include hypertension, diabetes, coronary heart disease, cerebral infarction, and chronic bronchitis. The source of the virus and the pathogenesis of this disease are still unconfirmed. No specific therapeutic drug has been found. The Chinese Government has initiated a level-1 public health response to prevent the spread of the disease. Meanwhile, it is also crucial to speed up the development of vaccines and drugs for treatment, which will enable us to defeat COVID-19 as soon as possible. World Health Organization (WHO) on March 11 declared COVID-19 a pandemic, pointing to the over 118,000 cases of the coronavirus illness in over 110 countries and territories around the world and the sustained risk of further global spread. A national state of emergency and quarantine have been declared in the USA and in the midst of the COVID-19 world pandemic [45-52], everything normal has been derailed. One obvious concern is the high risk of false positive results generated by unreliable, biased, or non-transparent algorithms. Another is “surveillance creep,” when surveillance developed for a limited purpose, such as fighting a pandemic or filming traffic violations, becomes used in ever more pervasive and permanent ways. How can we design the health surveillance and AI tools [53-60] needed to control COVID-19 and future pandemics so they don’t backfire or affect future wellbeing?
The potential route of COVID-19 infection: The bulk RNA-seq profiles [61-66] from two public databases including The Cancer Genome Atlas (TCGA) and Functional Annotation of The Mammalian Genome Cap Analysis of Gene Expression (FANTOM5 CAGE) dataset were collected (Figure 3). RNA-seq profiling data of 13 organ types with para-carcinoma normal tissues from TCGA and 14 organ types with normal tissues from FANTOM5 CAGE were analyzed in order to explore and validate the expression of ACE2 on the mucosa of oral cavity. Further, single-cell transcriptomes from an independent data generated in-house were used to identify and confirm the ACE2-expressing cell composition [67-70]. The results demonstrated that the ACE2 expressed on the mucosa of oral cavity. Interestingly, this receptor was highly enriched in epithelial cells of tongue. Preliminarily, those findings have explained the basic mechanism that the oral cavity is a potentially high risk for COVID-19 infectious susceptibility and provided a piece of evidence for the future prevention strategy in dental clinical practice as well as daily life.
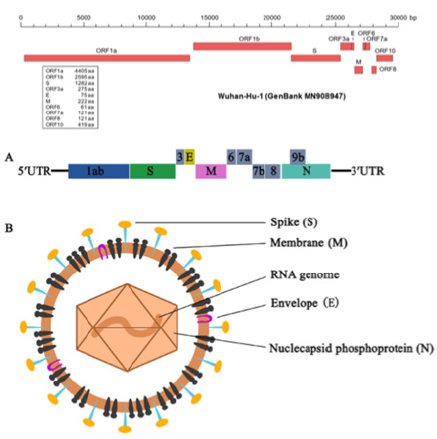
Genomic structure of COVID-19: The genome of COVID-19 [71-81] is consist of 6 major functional open reading frames (ORFs), including ORF1a/b, S, E, M, N and several other accessory genes like OR- F3b, OFR8 as shown in Figure 3. Replicase poly-proteins pp1a and pp1ab encoded by ORF1a/b would be proteolytic cleaved into 16 non-structural proteins (nsps) which are involved in the transcription and replication of the virus. In addition, the ORF3b encoded a completely novel short protein without exact function. The new ORF8 likely encodes a secreted protein formed by an alpha-helix, following with a beta-sheets containing six strands without known functional domain or motif similarly. The S gene of COVID-19 was less than 75% sequence identity to those of two CoVs, bat SARS-like CoVs (SL-CoVZXC21 and ZC45) and human SARS-CoV. And likewise, the spike glycoprotein encoded by the S genes of COVID-19 was longer than that of SARS-CoV. The spike protein, composed of S1 and S2 domain, was crucial to determine host tropism and transmission capacity through mediating receptor bind- ing and membrane fusion. Of which the S2 subunit of COVID-19 is highly con- served and has 99% identity with that of SARS-CoV. The receptor-binding do- main, commonly located in the C-terminal domain of S1 to directly contact the human receptor. Although, S1 domain of 2019-nCoV is only approximately 70% identity with SARS-CoV, homology modeling revealed that 2019-nCoV has a similar receptor-binding domain structure to that of SARS-CoV. Since the genomic sequences of COVID-19 obtained from different patients were extremely similar to each other, exhibiting more than 999% sequence identity, we could reasonably believe that COVID-19 originated from one source rather than a mosaic and could be detected relatively rapidly. However, mutations need to be constantly monitored when the virus is transmitting to an increasing number of individuals.
Figure 3: Genomic information on COVID-19 and its structure.
Computational identification of small interfering RNA targets in covid-19; theoretical predictions of the potential sirna targets in the virus genome: There are still some challenges that needed to be overcome for the clinic applications of siRNA, progresses have been made to solve the fundamental problems, such as off-target effects and effective delivery. For example, the position-specific chemical modification of siRNAs could can significantly reduce off targeting; safe and effective in vivo delivery systems have also been developed, such as nanoparticles, cationic lipids, antibodies, cholesterol, appetizes delivery strategies. Therefore, we hope that the above results (Table 1) would be useful in drug design and treatments against COVID-19.
Table 1:
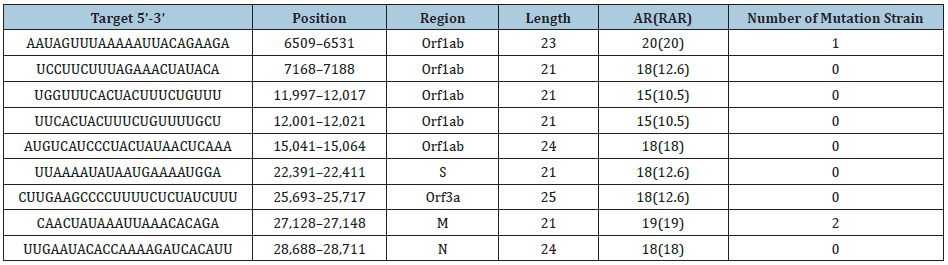
*The bold and underlined characters indicate the SNP found in different strains.
Figure 4: Machine learning including Active and Deep learning based on CT diagrams.
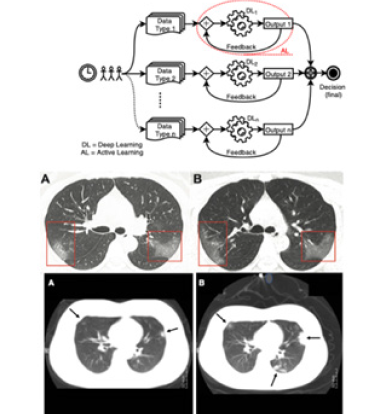
Machine learning as ai-driven tools: The novel coronavirus (COVID-19) outbreak, which was identified in late 2019, requires special attention because of its future epidemics and possible global threats. Beside clinical procedures and treatments, since Artificial Intelligence (AI) promises a new paradigm for healthcare, several different AI tools that are built upon Machine Learning (ML) algorithms are employed for analyzing data and decision-making processes. This means that AI-driven tools help identify COVID-19 outbreaks as well as forecast their nature of spread across the globe. However, unlike other healthcare issues, for COVID-19, to detect COVID-19, AI- driven tools are expected to have active learning-based cross-population train/test models that employs multi- tudinal and multi- modal data, which is the primary purpose of the paper. For time-series data, a schema of Active Learning (AL) model is provided. For better understanding, AL (in dotted red circle) is used with Deep Learning (DL) for all possible data types. In AL, expert’s feedback is used in parallel with the decisions from each data type. Since DL are data dependent, separate DLs are used for different data type. The final decision is made based on multi-tudinal and multimodal data (Figure 4). More often, AI-driven tools are limited to one data type. Decisions that are solely based on one data type (regardless of the data size) may be skewed away from the severity of coronavirus influence. In such a case, use of multi- tudinal and multimodal data can help support decision-making process with higher confidence. Since coronaviruses are enveloped viruses with a positive-sense single-stranded RNA genome and a nucleocapsid of helical symmetry, the most popularly used data for AI-driven tools mostly employ RNA sequences. Besides, Electronic Health Record (EHRs), Computerized Tomography (CT) scans, Chest X-rays (CRRs, Figures 5 & 6), and other data are considered and tested. Alibaba launched a new AI-based system to detect coronavirus infection via CT scans with an accuracy of up to 96%.
Figure 5: Expanded scheme for SEI/AR mathematical modeling.
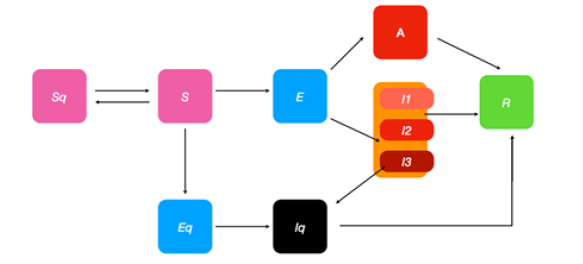
Figure 6: Physical distance invention simulation algorithms in infectious status.
Validation of statistical models on predictions for viruses capable of interspecies transmission mathematical model of infection kinetics and its analysis for COVID-19 Epidemic transmission: Based on the susceptible-exposed-infected/asymptomatic-removed (SEIAR) compartment model and the assumption that the infectious cases with symptoms occurred from free propagation without intervention, we estimate the basic reproduction number of COVID-19 according to the reported confirmed cases and suspected cases, as well as the theoretical estimated number of infected cases by other research teams, together with some epidemiological determinants learned from the severe acute respiratory syndrome (SARS). The modified SEIAR model is a classical epidemic model for the flows of people between five states: susceptible (S), exposed (E), infected (I), Asymptomatic(A), and recovery (R). Each of those variables represents the number of people in those groups. The relationship among the four groups is elucidated in Figure 7, where β1 is the prob- ability of S to E after I contacts S, γ1 is the probability of E to I, and γ2 is the probability of I to R. Since COVID-19 is also infectious in the incubation period, we introduced parameter β2 here to represent the probability of S to E after E contact S. We used the “susceptible-exposed-infected/asymptomatic-recovered” model to describe the prevalent characteristics of COVID-19. This is an ordinary differential equation model, described by the following equations:
Figure 7: Environmental mitigation control in the transmission.
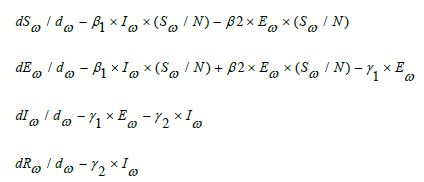
Among which, S(t), E(t), I(t), A(t), and R(t) represent the number of people in the group of the susceptible, the exposed, the infected and the recovered on the day t, respectively. N is the total number of possible contact people, which is assumed to be fixed and N=S+E+I/A+R.
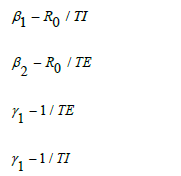
Parameters β1, β2, γ1 and γ2 were estimated according to the reference using the formula below: among which, R is the basic reproduction number, T1 is the time of infectious period, TE is the time of incubation period, and beta, beta, gamma, and gamma same meanings as in Figure 5.
Conceptualization of COVID-19 deposition to reduce transmission: Once an individual has been infected with COVID-19, viral particles accumulate in the lungs and upper respiratory tract. Droplets and aerosolized viral particles are expelled from the body through daily activities, such as coughing, sneezing, and talking, and non-routine events such as vomiting, and can spread to nearby surroundings and individuals. Viral particles, excreted from the mouth and nose, are often found on the hands and can be spread to commonly touched items such as computers, glasses, faucets, and countertops. There are currently no confirmed cases of fomite-to-human transmission, but viral particles have been found on abiotic environmental surfaces.
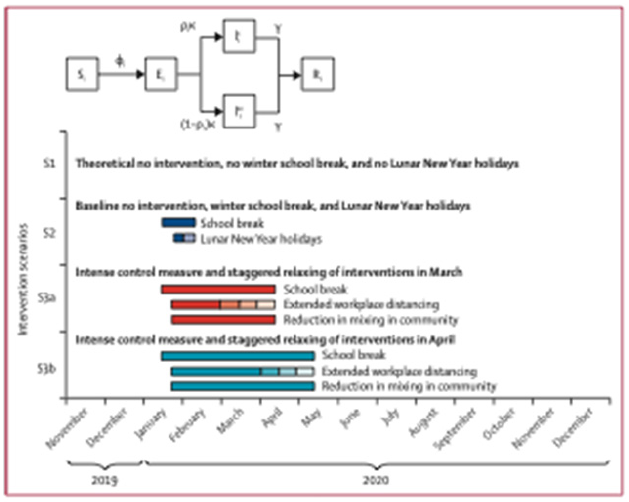
Details of the modeled physical distancing interventions: According to infection status, we divided the population into susceptible (S), exposed (E), infected (I), asymptomatic(A), and removed (R) individuals. An infected individual in an age group can be clinical (I) or subclinical (Ism), and prefers to the probability that an individual is symptomatic or clinical. The age-specific mixing patterns of individuals in age group i, C, alter their likelihood of being exposed to the virus given a certain number of infected individuals in the population. Younger individuals are more likely to be asymptomatic and less infectious, ie, subclinical. The transmission rate and α is the proportion of transmission that resulted from a subclinical individual. SEIAR= susceptible-exposed-infected-asymptomatic-removed. In addition to it, other factors could be influenced on the transmutability should be taken into consideration. Therefore, the estimates of the unreported cases between initial date and final date 2020, the basic reproduction Figure 8. The estimates of the unreported cases during designated period 2020, the basic number (R), and fitting results of the number of COVID-19 cases time series, which illustrated in Figure 9.
Figure 8: I value the simulation curve of the infected population and 2020 CDC coronavirus cases in South Korea.
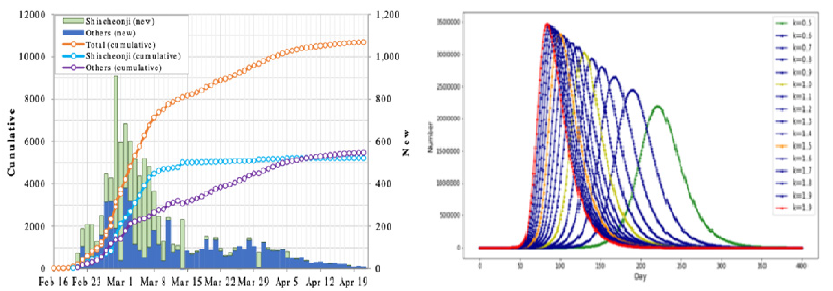
Figure 9: Differential equations utilized in the creation of mathematical models.

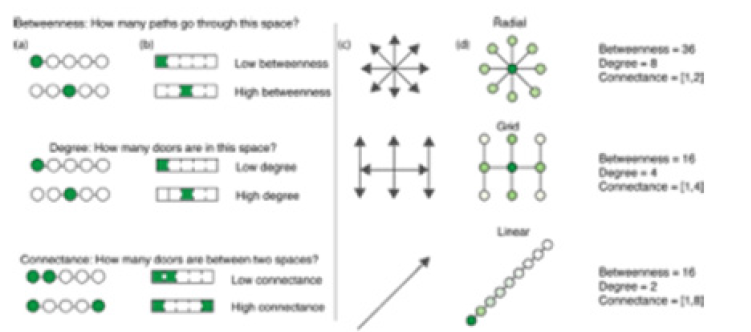
Control of mitigation efforts in the environmental transmission: The spread of COVID-19 is a rapidly developing situation, but there are steps that can be taken, inside and outside the host environment, to help prevent the spread of Coronavirus infection disease. Spatial connectivity, highlighting betweenness and connection of common room and door configurations as shown in Figure 7. Circles and lines follow the classic network representation. The rectangles follow the architectural translation of networks. Shaded areas correspond to a measure of betweenness (the number of shortest paths between all pairs of spaces that pass through a given space over the sum of all shortest paths between all pairs of spaces in the building), degree (the number of connections a space has to other spaces between any two spaces), and connection (the number of doors between any two spaces). The arrows represent possible directions of microbial spread as determined by the layout of the one. The circles represent the current knowledge of microbial spread based on microbial abundance through environmental factors as determined by layout. Darker colors represent higher microbial abundance, and lighter colors represent lower microbial abundance. The number of individuals who have contracted or have been exposed to S COVID-19 has been increasing dramatically. Over a decade of microbiology of the environmental research has been reviewed to provide the most up-to-date knowledge into the control and mediation of common pathogen exchange pathways and mechanisms in the parsimonious environment with as much specificity to COVID-19 as possible. We hope this information can help to inform the decisions and infection control mechanisms that are implemented by corporate entities, federal, state, county, and city governments, universities, school districts, places of worship, prisons, health care facilities, assisted living organizations, daycares, homeowners, and other building owners and occupants to reduce the potential for transmission through environmental transmission mediated pathways. This information is useful to corporate and public administrators and individuals responsible for building design and operation in their decision-making process about the degree and duration of social-distancing measures during viral epidemics and pandemics.
Predictive mathematical models of the covid-19 pandemic: Modeling studies have contributed vital insights into the COVID-19 pandemic, and will undoubtedly continue. Early models pointed to areas in which infection was likely widespread before large numbers of cases were detected; contributed to estimating the reproductive number, case fatality rate, and how long the virus had been circulating in a community; and helped to establish evidence that a significant amount of transmission occurs prior to symptom onset. As shown in Figure 8 mathematical models can be profoundly helpful tools to make public health decisions and ensure optimal use of re- sources to reduce the morbidity and mortality associated with the COVID-19 pandemic, but only if they are rigorously evaluated and valid and their projections are robust and reliable. Numerous mathematical models are being produced to forecast the future of coronavirus disease 2019 (COVID- 19) epidemics in the US and worldwide. These predictions have far-reaching consequences regarding how quickly and how strongly governments move to curb an epidemic. However, the primary and most effective use of epidemiological models is to estimate the relative effect of various interventions in reducing disease burden rather than to produce precise quantitative predictions about extent or duration of disease burdens.
Discussion
During the period of an epidemic when human-to-human transmission is established and reported case numbers are rising exponentially, nowcasting and forecasting are of crucial importance for public health planning and control domestically and internally. Our findings suggest that independent self-sustaining human-tohuman spread is already present in multiple major cities, many of which are global transport hubs with huge numbers of both inbound and outbound passengers, which caused pandemic now.
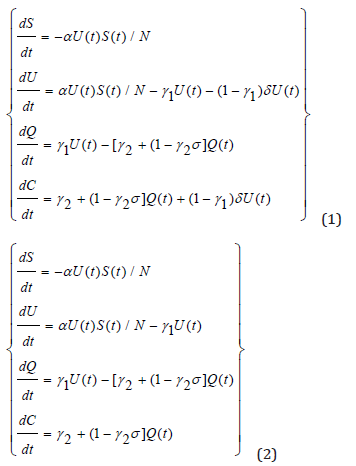
Conclusion
The implementation of more follow-up measures, including strict restrictions on people movement, accelerating the treatment of infected individuals, and clinical trials of new drugs would be required and the spread of COVID-19 pandemic will be effectively controlled, which the number of infected individuals will gradually decrease. It was expected that the epidemic would subside in early summer, and disappear gradually towards the late August. If the epidemic situation is not properly controlled, the peak of infect- ed number can be further increased and the peak time will be a little postponed. Current best evidence indicates that the most effective strategy to control the outbreak is the use of social distancing to break the chain of transmission.
Ethics
The study does not require ethical approval because the metaanalysis is based on published research and the original data are anonymous.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Maganga GD, Oppong S, Sarkodie Y, Vallo P, Silva Filho LV, et al. (2015) Evidence for an ancestral association of human coronavirus 229E with bats. J Virol 89(23): 11858-11870.
- Cottam EM, Whelband MC, Wileman T (2014) Coronavirus NSP6 restricts autophagosome expansion. Autophagy 10(18): 1426-1441.
- Angelini MM, Akhlaghpour M, Neuman BW, Buchmeier MJ (2013) Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce double-membrane vesicles. M Bio 4(4): e00524-e00513.
- Hulswit RJ, Haan CA, Bosch BJ (2016) Coronavirus spike protein and tropism changes. Adv Virus Res 96: 29-57.
- Gralinski LE, Ferris MT, Aylor DL, Whitmore AC, Green R, et al. (2015) Genome wide identification of SARS-CoV susceptibility loci using the collaborative cross. PLoS Genet 11: e1005504.
- Ge XY, Li JL, Yang XL, Chmura AA, Zhu G, et al. (2013) Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature 503(7477): 535-538.
- Frieman M, Ratia K, Johnston RE, Mesecar AD, Baric RS (2009) Severe acute respiratory syndrome coronavirus papain-like protease ubiquitin-like domain and catalytic domain regulate antagonism of IRF3 and NF-kappaB signaling. J Virol 83(13): 6689-6705.
- Du L, He Y, Zhou Y, Liu S, Zheng BJ, et al. (2009) The spike protein of SARS-CoV-a target for vaccine and therapeutic development. Nat Rev Microbiol 7(3): 226-236.
- Wilde AH, Falzarano D, Zevenhoven DJC, Beugeling C, Fett C, et al. (2017) Alisporivir inhibits MERS- and SARS-coronavirus replication in cell culture, but not SARS-coronavirus infection in a mouse model. Virus Res 228: 7-13.
- Angelini MM, Akhlaghpour M, Neuman BW, Buchmeier MJ (2013) Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce double-membrane vesicles. M Bio 4(4): e00524-e00513.
- Thornbrough JM, Jha BK, Yount B, Goldstein SA, Li Y, et al. (2016) Middle East respiratory syndrome coronavirus NS4b protein inhibits host RNase L activation. M Bio 7(4): e002587.
- Shirato K, Kawase M, Matsuyama S (2013) Middle East respiratory syndrome coronavirus infection mediated by the transmembrane serine protease TMPRSS2. J Virol 87(23): 12552-12561.
- Baric RS, Katze MG (2014) Cytokine systems approach demonstrates differences in innate and pro-inflammatory host responses between genetically distinct MERS-CoV isolates. BMC Genom 15(1): 1161.
- Rabouw HH, Langereis MA, Knaap RC, Dalebout TJ, Canton J, et al. (2016) Middle East respiratory coronavirus accessory protein 4a inhibits PKR-mediated antiviral stress responses. PLoS Pathog 12(10): e1005982.
- Lui PY, Wong LY, Fung CL, Siu KL, Yeung ML, et al. (2016) Middle East respiratory syndrome coronavirus M protein suppresses type I interferon expression through the inhibition of TBK1-dependent phosphorylation of IRF3. Emerg Microbes Infect 5(4): e39.
- Wilde AH, Jochmans D, Posthuma CC, Zevenhoven DJC, Nieuwkoop S, et al. (2014) Screening of an FDA-approved compound library identifies four small-molecule inhibitors of Middle East respiratory syndrome coronavirus replication in cell culture. Antimicrob Agents Chemother 58(8): 4875-4884.
- Wilde AH, Raj VS, Oudshoorn D, Bestebroer TM, Nieuwkoop S, et al. (2013b) MERS-coronavirus replication induces severe in vitro cytopathology and is strongly inhibited by cyclosporin A or interferon-alpha treatment. J Gen Virol 94(8): 1749-1760.
- Bailey EBA, Knaap RC, Johnson GG, Dalebout TJ, Ninaber DK, et al. (2014) Crystal structure of the Middle East respiratory syndrome coronavirus (MERS-CoV) papain-like protease bound to ubiquitin facilitates targeted disruption of deubiquitinating activity to demonstrate its role in innate immune suppression. J Biol Chem 289(50): 34667-34682.
- Brierley I, Dos Ramos FJ (2006) Programmed ribosomal frameshifting in HIV-1 and the SARS-CoV. Virus Res 119(1): 29-42.
- Digard P, Inglis SC (1989) Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell 57(4): 537-547.
- Brockway SM, Clay CT, Lu XT, Denison MR (2003) Characterization of the expression, intracellular localization, and replication complex association of the putative mouse hepatitis virus RNA-dependent RNA polymerase. J Virol 77(19): 10515-10527.
- Cruz JL, Sola I, Becares M, Alberca B, Plana J, et al. (2011) Coronavirus gene 7 counteracts host defenses and modulates virus virulence. PLoS Pathog 7(6): e1002090.
- Xinhua (2020) China’s CDC detects a large number of new coronaviruses in the South China seafood market in Wuhan. China.
- Huang C, Wang Y, Li X (2020) Clinical features of patients infected with novel coronavirus in Wuhan, China. Lancet 395(10223): 497-506.
- Rothe C, Schunk M, Sothmann P (2020) Transmission of 2019- nCoV infection from an asymptomatic contact in Germany. N Engl J Med 382(10): 970-971.
- Li Q, Guan X, Wu P (2020) Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med 382(13): 1199-1207.
- Wang C, Horby PW, Hayden FG, Gao GF (2020) A novel coronavirus outbreak of global health concern. Lancet 395(10223): 470-473.
- https://www.worldometers.info/coronavirus/
- Richman DD, Whitley RJ, Hayden FG (2016) Clinical Virology. ASM Press, USA.
- Chan Yeung M, Xu RH (2003) SARS: epidemiology. Respirology 8(1) S9-14.
- https://www.who.int/emergencies/mers-cov/en/.
- https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports
- Cheng ZJ, Shan J (2019) Novel coronavirus: where we are and what we know. Infection 48(52): 155-163.
- Chan JFW, Koi KW, Tse H, Jin DY, Yuen KY (2013) Interspecies transmission and emergence of novel viruses: lessons from bats and birds. Trends Microbiol 21(10): 544-555.
- King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (2012) Virus taxonomy, the ninth report of the international committee on taxonomy of viruses. Nucleic Acids Res 46(1): 810-814.
- Lau SKP (2013) Genetic characterization of beta-coronavirus lineage C viruses in bats reveals marked sequence divergence in the spike protein of Pipistrellus bat coronavirus HKU5 in Japanese Pipistrelle: Implications for the origin of the novel Middle East Respiratory Syndrome Coronavirus. J Virol 87(15): 8638-8650.
- Graham RL, Donaldson EF, Baric RS (2013) A decade after SARS: strategies for controlling emerging coronaviruses. Nat Rev Microbiol 11(12): 836-848.
- Ellis VA (2015) Local host specialization, host-switching, and dispersal shape the regional distributions of avian haemo- sporidian parasites. Proceedings of the National Academy of Sciences of the United States of America 112(36): 11294-11299.
- Hu B (2015) Bat origin of human coronaviruses. Virology Journal 12: 221.
- Anthony SJ (2012) Coronaviruses in bats from Mexico. Journal of General Virology 94(5): 1028-1038.
- Ann (2015) Non-random patterns in viral diversity. Nature Communications 6: 8147.
- Anthony (2017) Further evidence for bats as the evolutionary source of MERS coronavirus. M Bio 8(2): e00373-17.
- Azhar (2014) Evidence for camel-to-human transmission of MERS coronavirus. The New England Journal of Medicine 370(26): 2499-2505.
- Corman VM (2014a) Rooting the phylogenetic tree of middle east respiratory syndrome coronavirus by characterization of a conspecific virus from an African bat. Journal of Virology 88(19): 11297-11303.
- Consortium (2014) Reducing Pandemic Risk. USA.
- Irwin NR (2012) Complex patterns of host switching in New World arenaviruses. Molecular Ecology 21(16): 4137-4150.
- Jost L, Chao A, Chazdon RL (2011) Compositional similarity and beta diversity. USA.
- Karesh WB (2012) Ecology of zoonoses: natural and un-natural histories. Lancet 380(9857): 1936-1945.
- Keesing F (2010) Impacts of biodiversity on the emergence and transmission of infectious diseases. Nature 468: 647-652.
- Reusken CB (2010) Circulation of group 2 coronaviruses in a bat species common to urban areas in Western Europe. Vector Borne Zoonotic Dis 10(8): 785-791.
- Huang YW (2013) Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. M Bio 4(5): e00737-00813.
- Huynh J (2012) Evidence supporting a zoonotic origin of human coronavirus strain NL63. Journal of Virology 86(23): 12816-12825.
- Darriba D (2012) ModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9(8): 772.
- Rubin EJ, Baden LR, Morrissey S, Campion EW (2020) Medical journals and the 2019-nCoV outbreak. N Engl J Med 382(9): 866.
- Emerging understandings of 2019-nCoV. Lancet 395: 311
- Carlos WG, Dela Cruz CS, Cao B, Pasnick S, Jamil S (2020) Novel wuhan (2019-nCoV) coronavirus. Am J Respir Crit Care Med 201(4): P7-P8.
- Lei J, Li J, Li X, Qi X (2020) CT imaging of the 2019 Novel Coronavirus (2019-nCoV) pneumonia. Radiology 295(1): 18.
- Zhu N, Zhang D, Wang W, Li X, Yang B, et al. (2019) A novel coronavirus from patients with pneumonia in China. N Engl J Med 382(8): 727-733.
- Gralinski LE, Menachery VD (2020) Return of the Coronavirus: 2019- nCoV. Viruses 12(12): 135.
- Lin X, Gong Z, Xiao Z, Xiong J, Fan B (2020) Novel coronavirus pneumonia outbreak in 2019: computed tomographic findings in two cases. Korean J Radiol 21(3): 365-368.
- Subissi, Posthuma A, Collet JC, Zevenhoven DAE, Gorbalenya E, et al. (2014) One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc Natl Acad Sci 111(37): E3900-E3909.
- Kirchdoerfer RN, Ward AB (2019) Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat Commun 10(1): 2342.
- Lehmann KC, Gulyaeva A, Zevenhoven D, Janssen GMC, Ruben M, et al. (2015) Discovery of an essential nucleotidylating activity associated with a newly delineated conserved domain in the RNA polymerase-containing protein of all nidoviruses. Nucleic Acids Res 43(17): 8416-8434.
- Holshue ML, DeBolt C, Lindquist S, Lofy KH, Wiesman J, et al. (2020) Washington State 2019-nCoV Case investigation team, first case of 2019 novel coronavirus in the United States. N Engl J Med 382(10): 929-936.
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, et al. (2020) A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579(7798): 270-273.
- Chen C, Huang J, Cheng Z, Wu J, Chen S, et al. (2020) Favipiravir versus Arbidol for COVID-19: A randomized clinical trial. Med Rxiv, USA.
- Alenina N, Bader M (2019) ACE2 in brain physiology and pathophysiology: Evidence from transgenic animal models. Neurochem Res 44(6): 1323-1329.
- Chappell MC (2016) Biochemical evaluation of the renin-angiotensin system: the good, bad, and absolute? Am J Physiol Heart Circ Physiol 310(2): 137-152.
- Chappell MC, Marshall AC, Alzayadneh EM, Shaltout HA, Diz DI (2014) Update on the Angiotensin converting enzyme 2-Angiotensin (1-7)-MAS receptor axis: fetal programing, sex differences, and intracellular path- ways. Front Endocrinol (Lausanne) 4: 201–215.
- Chen D, Li X, Song Q, Hu C, Su F, et al. (2020) Hypokalemia and clinical implications in patients with Coronavirus Disease 2019 (COVID-19). MedRxiv.
- Chen Y, Liu Q, Guo D (2020) Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol 92(4): 418-423.
- Seah I, Agrawal R (2020) Can the coronavirus disease 2019 (COVID-19) affect the eyes? A review of coronaviruses and ocular implications in humans and animals. Ocul Immunol Inflamm 28(3): 391-395.
- Cascella M, Rajnik M, Cuomo A, Dulebohn SC, Di Napoli R (2020) Features, evaluation and treatment coronavirus (COVID-19). StatPearls. Stat Pearls
- Woo PCY, Huang Y, Lau SKP, Yuen KY (2010) Coronavirus genomics and bioinformatics analysis. Viruses 2(8): 1804-1820.
- Boheemen S, Graaf M, Lauber C, Bestebroer TM, Raj VS, et al. (2019) Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. M Bio 3(6). e00473e12.
- Yang D, Leibowitz JL (2015) The structure and functions of corona- virus genomic 30 and 50 ends. Virus Res 206: 120-133.
- Lu R, Zhao X, Li J, Niu P, Yang B, et al. (2020) Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395(10224): 565-574.
- Guo YR, Cao QD, Hong ZS, Tan YY, Chen SD, et al. (2020) The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) out-breakean update on the status. Mil Med Res 7(1): 1-10.
- Sola I, Almazan F, Zuniga S, Enjuanes L (2015) Continuous and discontinuous RNA synthesis in coronaviruses. Ann Rev Virol 2: 265-288.
- Chen N, Zhou M, Dong X, Qu J, Gong F, et al. (2020) Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet 395(10223): 507-513.
- Liu C, Zhou Q, Li Y, Garner LV, Watkins SP, et al. (2020) Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. ACS Cent Sci 6(3): 315-331.
© 2020 Hyunjo Kim. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






