- Submissions

Full Text
Cohesive Journal of Microbiology & Infectious Disease
General Overview of Recombinant DNA Technology Applications and Usage Areas in Clinical Microbiology
Gülnur Tarhan*
Department of Medical Microbiology, Turkey
*Corresponding author:Gülnur Tarhan, Department of Medical Microbiology, Turkey
Submission: September 27, 2019; Published: October 16, 2019

ISSN 2578-0190 Volume3 issues2
Abstract
Recombinant DNA technology is the desired modification of genetic material in-vivo. This technology involves inserting DNA fragments from various sources and transferring the desired DNA sequence with a vector. Manipulation in the genome of the organism can be accomplished by adding new genes or by limiting the activity of existing genes. Enzymatic digestion is used to obtain different DNA fragments. The DNA ligase enzyme is also used to insert DNA fragments into the vector. The vector is then transferred to the host cell. As the host cells proliferate, multiple copies of the vector and DNA fragment occur. In the last step, the cells carrying the transferred DNA sequence are selected and amplified. Today, most of the products used in the field of medicine, food, chemistry, biology and agriculture are produced with recombinant DNA technology. This technology has become the basis for more advanced methods that are in use today.iology
Keywords: Recombinant DNA; Usage area; Microbiology
Introduction
Recombinant DNA, which means combining two different DNA molecules, is a name given to molecular biology techniques commonly used for years [1]. Recombinant DNA technology and genetic engineering methods and concepts have been developed as a result of long studies on the basic structure of DNA molecule. The basic application steps used in this technology are shown in (Table 1) [2-4].
Table 1:Basic application step of recombinant DNA technology
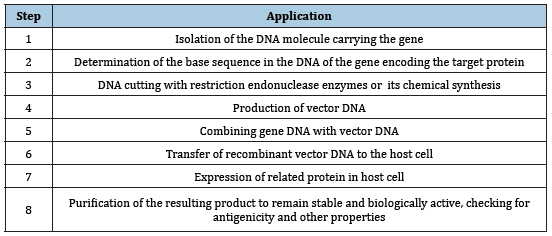
Detection of base sequences of nucleic acids (sequence analysis) was first performed for the RNA molecule. In 1964 alanine t-RNA was detected in yeast cells [5,6]. After this date, the chromosome sequence of single-strand RNA phage MS-2 was determined in 1975 [7]. In these years, the sequence analysis of the DNA molecule could not be performed due to the absence of specific enzymes. Since the deoxyribonuclease (DNase) enzymes cut the DNA molecule from non-specific regions, the heterogeneity of the resulting fragments made the analysis difficult. DNA sequence analysis was performed by the presence of restriction endonuclease enzymes that cut DNA from specific regions. These enzymes obtained from bacteria are used selectively for recombinant DNA technology. These are very specific deoxyribonuclease that are capable of recognizing and cutting double-stranded DNA from specific nucleotide sequences 4 to 6 bases in length [5,8]. The restriction endonuclease enzymes are called restriction fragments which are cut by the enzymes that are specific to the same DNA molecule, which can be obtained at any time using the specific enzyme. Since the molecular weights and sizes of the fragments obtained in this way are different from each other, it is possible to distinguish them using agarose gel electrophoresis. Each enzyme may produce sticky or blunt results at the cutting site. Such adhesive tip, as shown in the EcoR1 example; the end from a different DNA fragment cut by the EcoR1 enzyme may be joined to the sticky end and resulting in a recombinant DNA molecule [9,10]. Thermal transferase enzyme is used to make sticky ended DNA fragments. Thermal transferase is an enzyme that catalyzes the addition of deoxynucleotides to the 3end of DNA. It has catalytic activity in adding poly A or poly T to the adhesive ends. Adhesive ends generated by the terminal transferase enzyme are joined using the DNA ligase enzyme [11]. Obtaining restriction fragments facilitated the development of DNA nucleotide sequence methods. In 1977, Allan Maxam and Walter Gilbert determined the sequence of the SV40 genome. This technique is currently used as Maxam-Gilbert sequence analysis [12]. After that, Fred Sanger developed a new method using 2’-3 ’dideoxynucleosides, and DNA sequence analyzes became methods that could be completed in 1-2 weeks, unlike amino acid detection in proteins. In parallel with the development of DNA sequence analyzes, synthesis of oligonucleotides was also possible. Nucleotide sequences of 12-20 bases in length could be chemically synthesized in 1-2 days by this route [13,14].
The major innovation of recombinant DNA technology is that the gene fragment or the entire gene from an organism is transferred to a different organism and then cloned. With this method, the number of eukaryotic gene fragments in bacterial cells is increased and protein synthesis from these gene fragments may be possible with the addition of the necessary control elements. In gene cloning, intermediary molecules called vectors are utilized [3,4]. Vectors used for this purpose; Plasmid vectors, phage vectors, viral vectors, bacterial vectors are grouped into 4 types. Although genetic material exchange between different strains of bacteria has been known for many years, the functions and structures of plasmids have been started to be examined since the resistance to antibiotics started to emerge. Plasmids have the ability to impart resistance to antibiotics of bacterial cells, and to synthesize enterotoxin colistin and restriction endonucleases. Plasmids are double-stranded circular DNA molecules, ranging in size from 1 kb to 200kb. Plasmids capable of replicating independently of bacterial chromosomes and achieving high copy numbers can be used as vectors. The most important plasmids used for this purpose are; E. coli: Pbr322; P. aeruginosa: pRK2, pRSF100; S. aureus: pUB110, pC221; B. cereus: pBC16; B. subtulis pBS1; S. cerevisiae :2μ DNA plasmids [15,16].
Recombinant plasmids can transform bacterial cultures so that the foreign gene fragment they carry is cloned. The antibiotic resistance genes of vector plasmids contain restriction endonuclease recognition sites. When these regions are cut and the foreign DNA fragment is inserted, the resistance gene becomes inactive. The bacteria transformed with these plasmids in the presence of another antibiotic to which the plasmid is still resistant, selection of cells transformed with the recombinant plasmid can be made [2,17].
Plasmid pBR322 is the most widely used field to date. This plasmid has the ampicillin and tetracycline resistance gene and various restriction endonuclease enzyme recognition sites. Although plasmids are widely used as vectors, their small size can sometimes cause problems. They are generally used to clone DNA fragments of 4000 bases in length. As plasmids carrying large chromosomal DNA fragments proliferate, this DNA fragment may undergo deletion. Therefore, other vector systems are utilized to clone larger DNA fragments [18]. The vector used to clone DNA fragments about 15000 bases length was E. coli lambda phage. The lambda phage has a 50kb long, double-stranded linear DNA genome. It has a double-stranded linear DNA genome with 12 nucleotides length at both ends of DNA. There are 12 nucleotide long single strand complementary sequences at both ends of the DNA. These tips are sticky tips. In the host bacterial cell, the lambda phage becomes circular and replicates by means of adhesive tips. 60% of the genome of lambda phage is the part required for lytic infection in E. coli. 30% are not functional for lytic infection. This region is used to clone the foreign DNA fragment. In order for the recombinant phage to be stable, the size of the inserted DNA fragment and the size of the phage are important [19,20]. Cosmid vectors are used to clone DNA fragments of approximately 30,000-45,000 bases in length. Cosmid vectors consisted of combining the “cos” (sticky ends) regions of the lambda phage with the replication origin of the plasmids. These small vectors carry antibiotic resistance genes and restriction endonuclease recognition sites [21,22]. Animal viruses have been studied to develop animal vectors for eukaryotic cells.
As with other vectors, the most important consideration was the presence of one or more restriction endonuclease recognition sites in these vectors of appropriate size and enabling cloning. SV40 (Simian virus 40) is the first eukaryotic DNA virus used as cloning vector. In SV40, whose entire genetic sequence is known, almost all of these sequences are necessary for virus replication. The foreign DNA fragment can be inserted in place of the viral genes and the helper virus is needed for the virus to multiply. In general, late replicating genes with more active promoters than early genes are more suitable for cloning [2,23]. Another vector used for this purpose is retroviruses, single-stranded RNA viruses. Viral RNA becomes double-stranded proviral DNA with the reverse transcriptase enzyme, while the ends of the viral RNA (LTR, long terminal repeats) are incorporated into the cellular DNA. A foreign gene sequence that can be inserted between two LTRs is thus introduced into cellular DNA [24]. As bacterial vectors, Salmonella typhimurium, Mycobacterium bovis BCG strain, E. coli K12 strain are the most commonly used vectors in recombinant DNA technology studies. In addition, hybrid living viruses, chimeric nonreplicating pseudovirions have been introduced as vector systems in recent years. In addition, hybrid living viruses and chimeric nonreplicating pseudovirions have been introduced as vector systems in recent years [25].
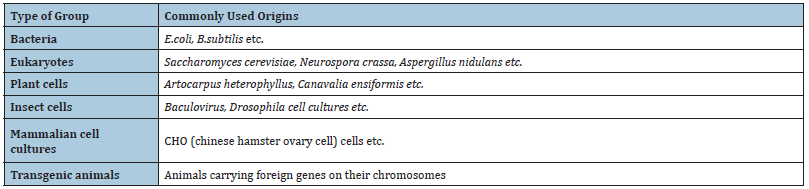
Recombinant protein is encoded by recombinant DNA, which has been cloned in a system that supports expression of the gene and translation of mRNA. The recombinant DNA, usually the cDNA sequence of the target protein, is designed to be under the control of a well characterized promoter and express the target protein within the chosen host cell to achieve high-level protein expression. Modification of the gene by recombinant DNA technology can lead to expression of a mutant protein or a large quantity of protein. Protein expression system is the key to the success of recombinant protein expression. Several factors need to be taken into consideration, including target protein property, intended application, protein yield. There are several expression systems for use. Different systems have different features and applications. Expression systems commonly used in this technology are given in (Table II) [26,27].
Table 2:Expression systems
Probe Preparation [2,3,28]: The double-stranded DNA strand is divided into two strands that are complementary to each other upon treatment with the base. Under suitable conditions, single strands are converted into double-stranded helix by hydrogen bonds made by DNA bases (Adenine-Thymine, Guanine-Cytosine). This property of the DNA molecule is based on DNA hybridization techniques. The gene fragment examined by hybridization methods is determined by the hybridization of the single stranded and labeled pure DNA probe with this complementary DNA sequence. DNA probes are prepared in three ways:
1. Genomic DNA from the cell nucleus
2. Complementary DNA (cDNA) synthesized from mRNA
3. Chemically synthesized DNA
The DNA probes may be all or part of the gene. And it can have all of the protein-coding or non-coding sequences. cDNA probes have only protein-coding sequences of the gene. The mRNA required to synthesize cDNA constitutes only 2% of cellular RNA. Therefore, the mRNA needs to be purified. mRNA is isolated by poly T cellulose affinity chromatography using the poly A sequence at the 3 end of the mRNA. Thereafter, the DNA-RNA heteroduplex from the mRNA by the reverse transcriptase enzyme from the retroviruses and then the double-stranded cDNA is synthesized and cloned by the appropriate vector. If the cDNA was obtained from a heterogeneous mRNA population, the gene-specific cDNA from the transformed bacterial cultures would have to be purified since it would also be heterogeneous. If plasmid is used as vector for this purpose, plasmid DNA is extracted from bacterial cells and it is absorbed to nitrous cellulose filters. After that, these filters are incubated with mRNA. Each filter only binds to the DNA specific mRNA it carries.
The DNA-RNA hybrids are then denatured, and the mRNA is removed from the filter. The protein obtained by in-vitro protein synthesis techniques is determined by immunological or biochemical methods. Thus, the cDNA copy of the examined gene is obtained. All of the cDNA clones of cellular mRNA are called the cDNA library. It is possible to obtain synthetic DNA strands using organic chemistry methods. However, due to the complexity of this method, it is only suitable for the synthesis of small sequences. There may be some benefits of this method. For example, A functional gene synthesis from the protein amino acid sequence is possible even if the nucleotide sequence of the gene is not known. mRNAs from eukaryotic cells can also be used as probes. Sometimes mRNA is very pure and high concentrations, such as globin mRNA. If mRNA is not pure and abundant, some problems arise in its use. mRNA isolation is difficult, and nuclease enzymes easily break down mRNA. In some cases, it may be necessary to use only RNA probe such as viral RNA, rRNA, tRNA.
Probe hybridization with complementary nucleotide sequences can only be demonstrated by a label attached to the prob. The most commonly used method should be labeled in radioisotope. Preferred radioactive markers for nucleic acid hybridization are 32P, 125I, 3H. The half-life of these isotopes is 14.3 days, 60.0 days and 12.35 years, respectively. In terms of high energy, 32P is the most preferred signal. Deoxyribonucleotides are available in 32P labeled forms. For 32P marking, the nick translation method is usually used.
In this method, in the double-stranded probe DNA, nickel is produced by the DNase enzyme. Single-stranded DNA breaks from the fractures continue using 5 polymer-3 nuclease activity of DNA polymerase I enzyme. The polymerase I activity of the enzyme synthesizes at fracture sites, while 32P-labeled deoxyribonucleotides are incorporated into DNA. And the probe DNA is marked as radioactive. Another method with high specific activity is oligolabelling. According to this method, the denatured probe DNA is double stranded again using hexadeoxynucleotide primers and DNA polymerase enzyme. One of the four nucleotides used during synthesis is used with the 32P tag. The filter is incubated with a specific labeled probe. The probe hybridizes with the base sequence which is complementary to it. Using autoradiography, this filter is incubated for 1-2 days with a special film. The radioactive signal allows us to localize the specific gene fragment. In addition, DNA or RNA probes can be labeled at the 5 and 3 ends using T4- polynucleotide kinase, terminal transferase enzymes. Factors such as the potential danger of radioisotopes and the short life span of the 32P isotope have recently increased their work on non-radioactive labeling methods. Non-radioactive marking can be of two types:
1. An enzyme can be bound to DNA or the DNA sample is modified by a method such as biotinylation. In the method of enzyme binding to DNA, the enzyme first binds to a positively charged polymer, and this polymer electrostatically adheres to the DNA. With this method, horseradish peroxidase and alkaline phosphatase enzymes can bind to DNA with high efficiency. Low molecular weight histone proteins bearing amino groups can be used as polymer. The color reaction obtained using the substrate of the bound enzyme is followed by hybridization to the probe nucleotide sequence of the probe.
2. Use of biotinylated nucleotides (e.g. Bio11-UTP) to label the DNA probe. The DNA is labeled with biotin by nick translation method. And biotinylated DNA is detected using avidin, which can bind biotin. As avidin is prepared based on an enzyme that catalyzes a colorimetric reaction, such as phosphatase peroxidase, the reaction of the color of the probe to the target nucleotide sequence is followed. Disadvantages of non-radioactive labelling methods: It’s not sensitive than labelling with 32P. If sensitivity limits are increased, cheap cost of testing and reduced detection time to as short as three hours makes non-Rayo active marking methods advantageous. Analysis of eukaryotic genes in the presence of suitable c DNA or mRNA probes can be performed without using the cloning method. The methods used for this purpose:
• Southern blot hybridization
• Northern blot hybridization
• Dot blot hybridization
• In-situ hybridization
• PCR (Polymerase chain Reaction)
In the method known as Southern blot and southern transfer; High molecular weight DNA is cutting using restriction endonuclease. The resulting DNA fragments are separated by agarose gel electrophoresis. After this step, the gel is placed on the nitrocellulose filter to ensure that the DNA bands in the gel pass to the filter through the appropriate buffer system. Nitrocellulose filter is dried at high temperature and immobilization of DNA probes takes place. The Southern Blot method, which is considered to be one of the most important methods of recombinant DNA technology with cloning techniques, was able to determine how many copies of a gene are present in a genome, the presence and number of specific restriction endonuclease cleavage sites in a gene. In the PCR technique which has been used in recent years; It is mainly based on in-vitro amplification of target DNA with the aid of specific deoxynucleotide primers and heat-resistant DNA polymerase (Taq polymerase or Tth polymerase) enzyme. With this method, a single DNA sequence can be reached to millions of numbers in as little as 2-3 hours and then identified by labeled probes. For this, Double stranded target DNA is added to the test medium together with buffer. Then, sufficient amount of deoxynucleotide tri phosphate (dATP, dGTP, dCTP, dTTP) specific oligonucleotide DNA primers and heat resistant Taq polymerase enzyme are added to the medium. After that, denaturation of the target DNA at 95oC, binding of primary DNA at 50oC and polymerization at 70-72oC were done.
Application Areas and Advantages of Recombinant DNA Technology in Clinical Microbiology
Recombinant DNA technology has opened new horizons in many fields of science such as medicine, pharmacy, biology, nutrition, biochemistry, microbiology, genetics, immunology, agriculture and environmental engineering. This technology is currently used in both basic and research studies. Small molecular weight compounds and macromolecules, which are naturally present in very low amounts, can be synthesized in large quantities. In addition, gene products could never be produced for mutagenesis or selection. The first product obtained in this way is pure human insulins prepared to avoid immunological side effects against insulin isolated from animal origins [29,30]. Human growth hormone and erythropoietin followed. In later stages:
• Colony stimulating factors used in bone marrow failure, some cancers and severe infectious diseases [31-33].
• Alpha, beta, gamma interferon and interleukins used in immune deficiencies due to T lymphocyte failure [2,34-36].
• Drugs called activase, tissue plasminogen activator for cardiovascular diseases and thromboembolic conditions [37].
• Recombinant factor used in the treatment of hemophilia in hematological VIIa [38].
• Various diagnostic agents that allow complete characterization and early monitoring of infectious and genetic diseases [33].
• Various organic chemicals and industrial products were produced [39].
• Antiviral and anticancer agents, immunoglobulins, enzymes of blood coagulation and vaccine production and treatment of primary metabolic insufficiencies [32,33,36].
• Combination of E. coli labile toxin with B subunit to increase mucosal immunomodulation [40].
• Purified B. pertussis components, antigenic repetitive sequences of HBsAg, Plasmodium, Salmonella recombinants expressing E. coli fimbria and toxin subunits are tested in animals [41,42].
• Recombinant obtained with the Shigella sonnei gene linked to S. typhi 21 a DNA, provided protection against shigellosis and typhoid [43].
• Vaccinia virus expressing HSVgD (glycoprotein D) produced specific neutralizing antibodies in rabbits [44].
• Intranasal immunization was achieved with recombinant adenoviruses that express rotavirus VP7 and HSVgD antigens [44].
• Recombinant BCG expressing Leishmania gp63 antigen produced protective cellular immunity [45].
• Clinical trials for recombinant BCG Osp A vaccine against Borrelia burgdorferi for Lyme disease were showed strong immune response [46].
• When the recombinant HIV-gp 120 envelope protein + influenza hemagglutinin + ISCOM was injected into BALB / C mice with mixing, the HIV-1 specific CTL response was initiated [47].
• Hepatitis A vaccine with immunostimulant influenza virosomes (IRIV) (influenzae hemagglutinin membrane protein + HAV antigen) reconstituted with hepatitis A virus antigens was tested and high antibody titers were obtained with a single dose [48].
• Various antigenic determinants of ETEC pilus, Brucella, V. cholerae, K. pneumoniae, H. influenzae, P. aeruginozas, S. aureus, L. pneumophila, ETEC, B. anthracis, C. diphtheriae, C. tetani toxin, influenza virus hemagglutinin The rabies virus glycoprotein and malaria genes were cloned [32,33,42].
Conclusion
In conclusion, since the first date of the introduction of recombinant DNA technology, significant advances have been made in the diagnosis, treatment and prevention of infectious diseases in clinical microbiology. Today, most of the products used in the field of medicine, food, chemistry, biology and agriculture are produced with recombinant DNA technology. This technology has become the basis for more advanced methods that are in use today.
References
- Harvey Lodish, Arnold Berk, S Lawrence Zipursky, Paul Matsudaira, David Baltimore, et al. (2000) Molecular Cell Biology (4th edn), WH Freeman, New York, USA.
- Siddiqui MA (1982) Recombinant DNA technology and its application to developmental biology. J Craniofac Genet Dev Biol 2(1): 75-92.
- Recombinant DNA (1992) Grolier Electronic Publishing 423-430.
- J D Watson, J Tooze, D T Kurtz (1983) Recombinant DNA - A Short Course, Scientific American Books 12(4): 187-187.
- Holley RW, Madison JT, Zamir A (1964) A new method for sequence determination of large oligonucleotides. Biochemical and Biophysical Research Communications 17(4): 389–394.
- Holley RW, Apgar J, Everett GA, Madison JT, Marquisee M, et al. (1965) Structure of a ribonucleic acid. Science 147(3664): 1462–1465.
- Fiers W, Contreras R, Duerinck F, Haegeman G, Iserentant D, et al. (1976) Complete nucleotide sequence of bacteriophage MS2 RNA: primary and secondary structure of the replicase gene. Nature 260(5551): 500–507.
- Holley RW, Apgar J, Merrill SH, Zubkoff PL (1961) Nucleotide and oligonucleotide compositions of the alanine-, valine-, and tyrosine-acceptor soluble ribonucleic acids of yeast. J Am Chem Soc 83(23): 4861–4862.
- Roberts RJ (2005) How restriction enzymes became the workhorses of molecular biology. Proc Natl Acad Sci 102(17): 5905–5908.
- Roberts RJ, Vincze T, Posfai J, Macelis D (2010) REBASE--a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res 38: D234–D236.
- Benedict CL, Gilfillan S, Thai TH, Kearney JF (2000) Terminal deoxynucleotidyl transferase and repertoire development. Immunol Rev 175: 150–157.
- Maxam AM, Gilbert W (1977) A new method for sequencing DNA. Proc Natl Acad Sci 74(2): 560–564.
- Sanger F, Coulson AR (1975) A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol 94(3): 441–448.
- Sanger F, Air GM, Barrell BG, Brown NL, Coulson AR, et al. (1977) Nucleotide sequence of bacteriophage phi X174 DNA. Nature 265(5596): 687–695.
- Khan K. H (2009) Vectors used in gene manipulation-a retrospective. Advanced Biotech Journal online pp1-8.
- Nora LC, Westmann CA, Santana LM, Alves LF, Monteiro LMO, et al. (2018) The art of vector engineering: towards the construction of next‐generation genetic tools. Microb Biotechnol 12(1): 125-147.
- Lu S (2003) Rapid Screening of Recombinant Plasmids. Methods Mol Biol 235: 169-174.
- Balbás P, Soberón X, Merino E, Zurita M, Lomeli H, et al. (1986) Plasmid vector pBR322 and its special-purpose derivatives--a review. Gene 50(1-3): 3-40.
- Friedman DI, Court DL (2001) Bacteriophage lambda: alive and well and still doing its thing. Curr Opin Microbiol 4(2): 201–207.
- Murphey KC (1998) Use of bacteriophage lambda recombination functions to promote gene replacement in Escherichia coli. J Bacteriol 180(8): 2063–2071.
- Wahl GM, Lewis KA, Ruiz JC, Rothenberg B, Zhao J, et al. (1987) Cosmid vectors for rapid genomic walking, restriction mapping, and gene transfer. Proc Natl Acad Sci U S A 84(8): 2160–2164.
- Choo KH, Filby G, Greco S, Lau YF, Kan YW (1986) Cosmid vectors for high efficiency DNA-mediated transformation and gene amplification in mammalian cells: studies with the human growth hormone gene. Gene 46(2-3): 277-286.
- Strayer D.S (1996) SV40 as an effective gene transfer vector in vivo. J Biol Chem 271(40): 24741-24746.
- Kurian KM, Watson CJ, Wyllie AH (2000) Retroviral vectors. Mol Pathol 53(4): 173–176.
- Davison J (2002) Towards safer vectors for the field release of recombinant bacteria. Environ Biosafety Res 1(1): 9-18.
- Lotfi H, Sheervalilou R, Zarghami N (2018) An update of the recombinant protein expression systems of Cyanovirin-N and challenges of preclinical development. Bioimpacts 8(2): 139–151.
- He Y, Wang K, Yan N (2014) The recombinant expression systems for structure determination of eukaryotic membrane proteins. Protein Cell 5(9): 658–672.
- Vasavirama K (2013) Molecular probes and their applications Review Article. Int J Life Sc Bt & Pharm Res 2(2): 32-44.
- Nossal GJ (1980) Human insulin through recombinant DNA technology. Med J Aust 2(6):295-296.
- Johnson IS (1983) Human insulin from recombinant DNA technology. Science 219(4585): 632-637.
- Rusthoven J (1991) The potential role of recombinant hematopoietic colony-stimulating factors in preventing infections in the immunocompromised host. Can J Infect Dis 2(2): 74-88.
- Gorbach SL (1978) Recombinant DNA: an infectious disease perspective. J Infect Dis 137(5): 615-623.
- Cederbaum SD, Fareed GC, Lovett MA, Shapiro LJ (1984) Recombinant DNA in medicine. West J Med 141(2): 210-222.
- Arbabi M, Alasti F, Sanati MH, Hosseini S, Deldar A, et al. (2003) Cloning and expression of human gamma-interferon cDNA in E. coli. Iranian Journal of Biotechnology 1(2): 87-94.
- Akdis M, Aab A, Altunbulakli C, Azkur K, Costa RA, et al. (2016) Interleukins (from IL-1 to IL-38), interferons, transforming growth factor β, and TNF-α: Receptors, functions, and roles in diseases. J Allergy Clin Immunol 138(4): 984-1010.
- Bhopale GM, Nanda RK (2005) Recombinant DNA expression products for human therapeutic use. Current Science 89(4): 614-622.
- Grossbard EB (1987) Recombinant tissue plasminogen activator: a brief review. Pharm Res 4 (5): 375-378.
- Lyseng-Williamson KA, Plosker GL (2007) Recombinant factor VIIa (eptacog alfa): a pharmacoeconomic review of its use in haemophilia in patients with inhibitors to clotting factors VIII or IX. Pharmacoeconomics 25(12): 1007-1029.
- Jones MD, Fayerman JT (1987) Industrial applications of recombinant DNA technology. J Chem Educ 64(4):337.
- Martin M, Hajishengallis G, Metzger DJ, Michalek SM, Connell TD, et al. (2001) Recombinant antigen-enterotoxin A2/B chimeric mucosal immunogens differentially enhance antibody responses and B7-dependent costimulation of CD4(+) T cells. Infect Immun 69(1): 252-261.
- Liew FY (1990) Biotechnology of vaccine development. Biotechnol Genet Eng Rev 8: 53-95.
- Nascimento IP, Leite LCC (2012) Recombinant vaccines and the development of new vaccine strategies. Braz J Med Biol Res 45(12): 1102–1111.
- Wu Y, Chakravarty S, Li M, Wai TT, Hoffman SL, et al. (2017) Development of a live attenuated bivalent oral vaccine against Shigella sonnei Shigellosis and typhoid fever. J Infect Dis 215(2): 259-268.
- Bernstein DI (2000) Effect of route of vaccination with vaccinia virus expressing HSV-2 glycoprotein D on protection from genital HSV-2 infection. Vaccine 18(14): 1351-1358.
- Afkhami S, Yao Y, Xing Z (2016) Methods and clinical development of adenovirus-vectored vaccines against mucosal pathogens. Mol Ther Methods Clin Dev 27(3): 16030.
- Sikand V K, Halsey N, Krause PJ, Sood S K, et al. (2001) Safety and immunogenicity of a recombinant Borrelia burgdorferi outer surface protein A vaccine against lyme disease in healthy children and adolescents: a randomized controlled trial. Pediatrics 108(1): 123-128.
- Xu W, Zhao Q, Xing L, Lin Z (2016) Recombinant production of influenza hemagglutinin and HIV-1 GP120 antigenic peptides using a cleavable self-aggregating tag. Sci Rep 3(6): 35430.
- Glück R, Mischler R, Brantschen S, Just M, Althaus B, et al. (1992) Immunopotentiating reconstituted influenza virus virosome vaccine delivery system for immunization against hepatitis A. J Clin Invest 90(6): 2491-2495.
© 2019 Gülnur Tarhan. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






