- Submissions

Full Text
Cohesive Journal of Microbiology & Infectious Disease
Molecular Dıagnostic Tests Used in the Dıagnosis of Tuberculosis
Gülnur Tarhan*
Department of Medical Microbiology, Adıyaman University, Turkey
*Corresponding author:Gülnur Tarhan, Faculty of Medicine, Department of Medical Microbiology, Adıyaman University, Turkey
Submission: July 30, 2018; Published: September 11, 2018

ISSN 2578-0190 Volume2 Issue2
Abstract
The definitive diagnosis of tuberculosis (TB) is dependent on the isolation, identification and drug susceptibility testing of the causal agent Mycobacterium tuberculosis by cultivation. Smear microscopy has poor sensitivity and culture is slow to yield results. The resurgence of tuberculosis worldwide has been accompanied by an increase in the incidence of multidrug-resistant (MDR) tuberculosis on all continents. At the same time, a number of other nontuberculous mycobacterial (NTM) species are emerging as causes of disease. A quick and correct diagnosis of symptomatic tuberculosis is critical for the control of this serious disease. The nucleic acid amplification techniques (NAATs) and other molecular biology methods (i .e. DNA hybridization, DNA sequencing, etc.) are essential in today’s laboratory practices for detection and characterization of mycobacteria. The use of NAATs in the routine detection of mycobacteria allowed a fast and accurate detection of the Mycobacterium species within 24 hours. The methods are widely used for the identification of mycobacteria, detection of the mutations in the resistance genes as well as the molecular epidemiological studies. The availability of new kits, and accumulated experience with nucleic acid amplification techniques for M. tuberculosis detection in most laboratories, have yielded improved sensitivity and specificity of these tests.
Keywords: Tuberculosis; Diagnosis; Serological tests
Introduction
Tuberculosis (TB), caused by Mycobacterium tuberculosis, is an old and serious infectious disease in humans, and it is still a major public health problem worldwide. It is estimated that nearly 1 billion people will be newly infected with TB between 2000 and 2020 and, furthermore, two hundred million people will develop disease and 35 million will die from TB within this period [1-4]. The definitive diagnosis of tuberculosis is dependent on the isolation, identification and drug susceptibility testing of the causal agent Mycobacterium tuberculosis by cultivation. Smear microscopy has poor sensitivity and culture is slow to yield results. A quick and correct diagnosis of symptomatic tuberculosis is critical for the control of this serious disease [5-8]. The resurgence of tuberculosis worldwide has been accompanied by an increase in the incidence of multidrug-resistant (MDR) tuberculosis on all continents. At the same time, a number of other nontuberculous mycobacterial (NTM) species are emerging as causes of disease [9-17]. The late 1990’s have brought significant changes to clinical mycobacteriology with the introduction of broth-based cultivation and molecular biological methods. Molecular diagnostic tools for TB have evolved quickly with new innovations which can provide unprecedented opportunities for the rapid, sensitive and specific diagnosis of M. tuberculosis in clinical specimens and the status of its drug sensitivity. However, the value of any new method can be estimated only on the basis of comparisons with conventional techniques currently available in clinical laboratories [18-24].
The nucleic acid amplification techniques (NAATs) and other molecular biology methods (i.e., DNA hybridization, DNA sequencing, etc.) are essential in today’s laboratory practices for detection and characterization of mycobacteria. NAATs are a molecular system that detects small amounts of genetic material from the microorganism (DNA or RNA). The nucleic acid must be extracted from the clinical specimen and afterwards amplified by PCR [21,23]. Using the nucleic acid amplification techniques, in theory, a single mycobacterial DNA can be amplified with an enzymatic reaction to the detectable amount in a very short period of time [22-24]. The use of NAATs in the routine detection of mycobacteria allowed a fast and accurate detection of the Mycobacterium species within 24 hours. The methods are widely used for the identification of mycobacteria, detection of the mutations in the resistance genes as well as the molecular epidemiological studies. The insertion element IS 6110 and the 16S rDNA are the most common targets used. Other regions used for amplification include the prob gene encoding the b-subunit of the RNA polymerase, the gene coding for the 32kD protein, the rec A gene, the hsp65 gene, the DNAJ gene, the sod A gene and the 16S-23S rRNA internal transcriber spacer [22-30]. There are many nucleic acid amplification techniques as commercial or in-house prepared kits for M. tuberculosis from clinical samples. The availability of new kits, and accumulated experience with nucleic acid amplification techniques for M. tuberculosis detection in most laboratories, have yielded improved sensitivity and specificity of these tests.
Commercially available NAATs
Cobas amplicor MTB test: The Cobas Amplicor MTB Test (Roche Molecular Systems, Basel, Switzerland) is a single unit combining five instruments (automated pipettor, incubator, thermal cycler, wash station and photometer), which enable automated amplification and detection of the M. tuberculosis complex. A 584 bp fragment of the 16S ribosomal RNA gene, comprising a speciesspecific region flanked by genus-specific sequences, is amplified using biotinylated primers [31-35]. The test includes four steps: specimen preparation, PCR amplification, hybridization and detection. In brief, specimens are liquefied and decontaminated with NALC-NaOH. A portion of 50μl of the processed specimen is added to the amplification mixture in amplification tubes containing Taq polymerase, biotinylated primers and abundant dNTPs including deoxyadenosine, deoxyguanosine, deoxycytidine and deoxyuridine (dUTP) in place of deoxythymidine. The amplification process includes denaturation of the double stranded DNA, annealing of the primers and extension of the amplicon sequence, which occur at different temperatures. The procedure is repeated for the required number of cycles, and consequently the copies of the original DNA sequence increase exponentially. Further, after hybridization of M. tuberculosis-specific DNA probe, the detection is accomplished by a colorimetric reaction measured with a photometer at wavelength of 660 nm. Absorbance values ≥0.35 are scored positive. The detection of the specific amplification product is performed by adding an avidin-enzyme conjugate and a chromogenic substrate [31,32]. The amplification and detection steps are carried out automatically by the Cobas Amplicor instrument. The method is approved by FDA, USA for testing smear-positive respiratory samples. It includes an internal control, composed of synthetic DNA characterized by identical annealing sequences as the mycobacterial target; when this is not amplified, it signals the presence of inhibitors. The detection of M. tuberculosis complex DNA can also be carried out without the Cobas instrument, using a manual kit that, however, does not include an internal control. Other Amplicor kits are available for detection of Mycobacterium avium and Mycobacterium intra cellular DNA in clinical samples. From the literature review, specificity is close to 100 % while sensitivity ranges from 90% to 100% in smear-positive samples and from 50 % to 95.9 % in smear negative ones [31-43].
Amplified Mycobacterium Tuberculosis Direct test (AMTD)
AMTD is an isothermal (42 °C) transcriptase-mediated amplification system by developed Gen Probe (San Diego, CA, USA). This system is amplifying rRNA (16S rRNA) by DNA intermediates. Briefly, the promoter-primer binds to the target rRNA and the reverse transcriptase enzyme creates DNA copy of the target. rRNA is degraded from the RNA-DNA duplex and the primer 2 anneals to the DNA and new DNA is made. Subsequently DNA-directed RNA polymerase transcribes RNA amplicons from the DNA template. New synthetised amplicons re-enter the TMA process, and repeated replication cycles produce a billion-fold amount of RNA amplicons. The amplicon products are detected with an acridinium ester-labelled DNA probe in a hybridization assay and the results are read by the luminometer [44,45]. The commercial TMA assay (AMTD2, Gen Probe) differs from the Cobas Amplicor test in some respects (Table 1).
Table 1:Differencies of the Cobas Amplicor and Amplified Mycobacterium Tuberculosis Direct(AMTD2) assays.
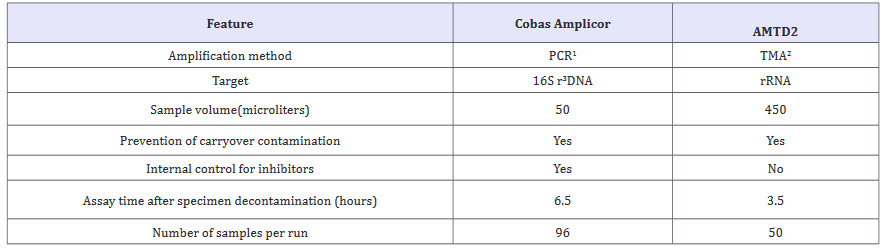
1 PCR, polymerase chain reaction; 2TMA, transcription-mediated amplification; 3Ribosomal.
Firstly, thousands of copies of the target rRNA are present in mycobacterial cells compared to 10 to 20 copies of target DNA used in the PCR assay. Thermal-cyclers are not needed and the whole amplification step is carried out on a heating block at 42 °C. The turnaround time is 2.5 hours. No internal control is provided in the kit to monitor the presence of inhibitors. The method is approved by FDA, USA for testing smear-positive and smear-negative respiratory samples. The overall sensitivity for respiratory specimens was found in the range between 90.9% and 95.2% and the specificity between 97.6% and 100% [44-49].
BD Probe Tec ET
The BD Probe Tec ET (Becton Dickinson, Sparks, MD) is the Probe Tec ET DTB (Becton Dickinson) test based on the strand displacement amplification (SDA). In the initial phase (target amplification), amplification is started by two pairs of primers complementary to contiguous sequences delimiting the target. The elongation of the upstream primer, also named bumper, determines the displacement of the simultaneously elongating downstream primer and finally releases the produced amplicon. A restriction site, present in the downstream primer, will also be present in the released amplicon. In the exponential amplification phase, a new primer anneals to the amplicon and, following digestion by the restriction enzyme, the upstream fragment acts as bumper and displaces the downstream fragment [50,51]. It is an automated isothermal method characterized by simultaneous DNA amplification and real-time fluorometric detection of the amplicons. An internal control to detect the presence of inhibiting substances is included in each run. The test performance time is approximately 4 hours after specimen preparation. The system is not yet approved by the US FDA. Kits are also available for the amplification of nucleic acids of M.avium,M. intracellular and Mycobacterium kansasii . The literature reports a rate of sensitivity ranging from 98.5 % to 100 % for smear positive samples and very variable (0.33%-100%) for smear-negative ones [50-54].
Genotype mycobacteria direct assay
This method includes RNA isolation, isothermal amplification and detection by reverse hybridization. It is based on a DNA strip technology, and in addition to M. tuberculosis complex it permits simultaneous detection of M. avium, M. intracellular, M. kansasii and M. malmoense (Hain Life science, Nehren, Germany). This novel assay is based on the nucleic acid sequence-based amplification (NASBA) applied to DNA strip technology. According to the manufacturer, the assay has three steps. The first step consists of isolation of 23S rRNA, the second step includes amplification of RNA by NASBA method, and the third step involves the reverse hybridization of the amplified products on membrane strips using an automated system. The assay has the ability for simultaneous detection of M. avium, M. intracellular, M. kansasii, M. malmoense and MTBC. Isolation of highly specific RNA is achieved by the use of the “magnetic bead capturing” method. This assay is useful, reliable and rapid, with sensitivity and specificity of 92% and 100%, respectively.
LCx MTBC assay
Ligase chain reaction (LCx) (Abbott Laboratories Diagnostic Division, Chicago, USA) is a method based on DNA amplification. The assay uses the ligase chain reaction for amplification of a target sequence within the chromosomal gene that codes for protein antigen b, which is specific for members of the MTBC. Two primers attach to each strand leaving a gap in between, which is filled by the action of DNA polymerase and the primers are linked together by ligase. The first pair of oligonucleotides acts as a template for new complementary oligonucleotides. Detection of the amplicons is carried out by microparticle enzyme assay with the LCx fluorometric analyzer. The LCx MTB assay (ABBOTT LCx probe system) has gone through various modifications [42,57]. This test has no internal control and the main shortcomings have been with inhibitory susbstances and lack of sensitivity. The overall sensitivity and specificity of the assay was 74% and 98%, respectively. For smear-positive samples the sensitivity reached 100%, but for smear-negative it was only 57%. In a multicenter evaluation of Amplicor and LCx, the sensitivity of both methods was significantly better when only respiratory specimens were considered (78% and 88%, respectively). When non-respiratory samples were used, the sensitivity was reduced to 59% for Amlicor and 65% for LCx [57- 60].
Real time PCR
Real-time PCR systems are the most common used in routine mycobacteriology laboratories. During the laboratory study, these systems allow real-time monitoring of a DNA amplification reaction by measuring an accumulating fluorescence signal. Realtime PCR provided improved sensitivity and specificity, reducing turnaround time and avoiding the use of ethidium bromide-stained gels. Different real-time instruments are now available in the market. In routine laboratory, Real-time PCR detection technology has been widely evaluated pulmonary and extrapulmonary samples for TB. The majority of real-time PCR methods reported to date for mycobacteria focus on detection of the Mycobacterium tuberculosis complex. Several publications address the detection of Mycobacteria at the genus level. The risk of contamination is considerably less with real-time PCR compared to conventional PCR, but it still can occur. Specimen to specimen contamination has become a greater challenge than amplified product contamination. The most obvious situation where specimen-to-specimen contamination can occur is with the transfer of specimen to the PCR vessel or to the DNA extraction tube.
References
- World Health Organisation Report (2006) Global tuberculosis control: surveillance, planning, financing. World Health Organization (WHO/ HTM/TB/2006.362), Geneva, Switzerland.
- World Health Organisation (2009) Global Tuberculosis Control; A short update to the 2009 report. 2009, World Health Organisation, Geneva, Switzerland.
- European Center Diseases Control (ECDC) (2010) Tuberculosis surveillance in Europe. European Centre for Disease Prevention and Control: Stockholm, Sweden.
- World Health Organisation (2006) Stop TB Strategy. Geneva, Switzerland
- Metchock BG, Nolte FS, Wallace RJ (1999) Mycobacterium, in Manual of clinical microbiology. In: PR Murray, (7th edn), ASM Press: Washington DC, USA, p. 399-437.
- Hale YM, Pfyffer GE and Salfinger M (2001) Laboratory diagnosis of mycobacterial infections: New tools and lessons learned. Clin Infect Dis 33: 834-846.
- Frieden TR, Sterling TR, Munsiff SS (2003) Tuberculosis. Lancet 13; 362 (9387): 887-899.
- Amdekar Y (2005) Tuberculosis - persistent threat to human health. Indian J Pediatr 72: 333-338.
- Manangan LP, Jarvis WR (2000) Preventing multidrug-resistant tuberculosis anderrors in tuberculosis treatment around the globe. Chest 117(3): 620-623.
- Grange JM, Zumla A (2000) The global emergency of tuberculosis: what is the cause? J R Soc Health 122: 78-81.
- Couvin D, Rastogi N (2015) Tuberculosis-A global emergency: Tools and methods to monitor, understand, and control the epidemic with specific example of the Beijing lineage. Tuberculosis 177-189.
- Raviglione MC, Snider DE Jr, Kochi A (1995) Global epidemiology of tuberculosis: Morbidity and mortality of a worldwide epidemic. JAMA 273(3): 220-226.
- Dye C, Scheele S, Dolin P, Pathania V, Raviglione MC (1999) Consensus statement. Global burden of tuberculosis: Estimated incidence, prevalence, and mortality by country. WHO Global Surveillance and Monitoring project. JAMA 282: 677-686.
- Anderson RM (1999) The pandemic of antibiotic resistance. Nat Med 5(2): 147-149.
- World Health Organization (2010) Multidrug and extensively drugresistant TB (M/XDR-TB) (2010) Global Report on Surveillance and Response, Geneva,15.
- World Health Organization (2010) The Global Plan to STOP TB 2011- 2015. Switzerland, Geneva.
- Sharma SK, Mohan A (2004) Multidrug-resi resistant tuberculosis. Indian J Med Res 120(4): 354-376
- Salfinger M, Pfyffer GE (1994) The new diagnostic mycobacteriology laboratory. Eur J Clin Microbiol Infect Dis 13(11): 961-979.
- Centers for Disease Control and Prevention (CDC) (2005) Trends in tuberculosis-United States. MMWR Morb Mortal Wkly Rep 55(11): 305- 308.
- Woods GL (2001) Molecular techniques in mycobacterial detection. Arch Pathol Lab Med 125 (1): 122-126.
- Shinnick TM, Good RC (1995) Diagnostic mycobacteriology laboratory practices, Clin Infect Dis 21(2): 291-299.
- Newton CR (1995) Setting up a PCR laboratory. In Newton CR (Ed.), PCR: Essential data, Online Wiley, New York, USA, p. 216.
- Engström A (2016) Fighting an old disease with modern tools: Characteristics and molecular detection methods of drug-resistant Mycobacterium tuberculosis. Infect Dis 48(1): 1-17.
- Brisson-Noel A, Aznar C, Chureau C, Nguyen S, Pierre C, et al. (1991) Diagnosis of tuberculosis by DNA amplification in clinical practice evaluation. Lancet 338(8763): 364-366.
- Soini H, Musser JM (2001) Molecular diagnosis of mycobacteria. Clin Chem 47(5): 809-814.
- Soini H, Viljanen MK (1997) Gene amplification in the diagnosis of mycobacterial infections. APMIS 105(5): 345-353.
- Shankar P, Manjunath N, Lakshmi R, Aditi B, Seth P, et al. (1990) Identification of Mycobacterium tuberculosis by polymerase chain reaction. Lancet 335(8686): 423.
- Thierry D, Brisson-Noël A, Vincent-Lévy-Frébault V, Nguyen S, Guesdon JL, et al. (1990) Characterization of a Mycobacterium tuberculosis insertion sequence, IS6110, and its application in diagnosis. J Clin Microbiol 28(12): 2668-2673.
- Thierry D, Cave MD, Eisenach KD, Crawford JT, Bates JH, et al. (1990) IS6110, an IS-like element of Mycobacterium tuberculosis complex. Nucleic Acids Res 18(1): 188.
- Kox LF, van Leeuwen J, Knijper S, Jansen HM, Kolk AH (1995) PCR assay based on DNA coding for 16S rRNA for detection and identification of mycobacteria in clinical samples. J Clin Microbiol 33(12): 3225-3233.
- Reischl U, Lehn N, Wolf H, Naumann L (1998) Clinical evaluation of the automated COBAS AMPLICORMTB assay for testing respiratory and nonrespiratory specimens. J Clin Microbiol 36(10): 2853-2860.
- Fegou E, Jelastopulu E, Sevdali M, Anastassioua ED, Dimitracopoulos G (2005) Sensitivity of the Cobas Amplicor system for detection of Mycobacterium tuberculosis in respiratory and extrapulmonary specimens. Clinical Microbiology Infection 11(7): 593-596.
- Bogard M, Vincelette J, Antinozzi R, Alonso R, Fenner T, et al. (2001) Multicenter study of a commercial, automated polymerase chain reaction system for the rapid detection of Mycobacterium tuberculosis in respiratory specimens in routine clinical practice. Eur J Clin Microbiol Infect Dis 20(10): 724-731.
- Levidiotou S, Vrioni G, Galanakis E, Gesouli E, Pappa C, et al. (2003) Four-year experience of use of the Cobas Amplicor system for rapid detection of Mycobacterium tuberculosis complex in respiratory and nonrespiratory specimens in Greece. Eur J Clin Microbiol Infect Dis 22(6): 349-356.
- Burggraf S, ReischlU, Malik N, Bollwein M, Naumann L, et al. (2005) Comparison of an internally controlled, large-volume Light Cycler assay for detection of Mycobacterium tuberculosis in clinical samples with the COBAS AMPLICOR assay. J Clin Microbiol 43(4): 1564-1569.
- Gomez-Pastrana D, Torronteras R, Caro P, Anguita ML, López-Barrio AM, et al. (2001) Comparison of amplicor, in-house polymerase chain reaction, and conventional culture for the diagnosis of tuberculosis in children. Clin Infect Dis 32(1): 17-22.
- Centers for Disease Control and Prevention (CDC) (2000) Notice to readers: Update: Nucleic acid amplification tests for tuberculosis. MMWR Morb Mortal Wkly Rep 49(26): 593-594.
- D´Amato RF, Wallman AA, Hochstein LH, Colaninno PM, Scardamaglia M, et al. (1995) Rapid diagnosis of pulmonary tuberculosis using Roche AMPLICOR Mycobacterium tuberculosis PCR test. J Clin Microbiol 33(7): 18321834.
- D´Amato RF, Hochstein LH, Colaninno PM, Scardamaglia M, Kim K, et al. (1996) Application of the Roche Amplicor Mycobacterium tuberculosis (PCR) test to specimens other than respiratory secretions. Diagn Microbiol Infect Dis 24(1): 15-17.
- Noordhoek GT, Kolk AH, Bjune G, Catty D, Dale JW, et al. (1994) Sensitivity and specificity of PCR for detection of Mycobacterium tuberculosis: a blind comparison study among seven laboratories. J Clin Microbiol 32(2): 277-284.
- Rimek D, Tyagi S, Kappe R (2002) Performance of an IS6110-based PCR assay and the Cobas Amplicor MTB PCR system for detection of Mycobacterium tuberculosis complex DNA in human lymph node samples. J Clin Microbiol 40(8): 3089-3092.
- Daniel TM (1990) The rapid diagnosis of tuberculosis: A selective review. J Lab Clin Med 116(3): 277-282.
- (1990) Diagnostic standards and classification of tuberculosis. Am Rev Respir Dis 142(3): 725-735.
- Coll P, Garrigó M, Moreno C, Marti N (2003) Routine use of Gen-Probe Amplified Mycobacterium tuberculosis Direct (MTD) Test for detection of Mycobacterium tuberculosis with smear-positive and smear-negative specimens. Int J Tuberc Lung Dis 7(9): 886-891.
- Bodmer T, Möckl E, Mühlemann K, Matter L (1996) Improved performance of Gen-Probe Amplified Mycobacterium tuberculosis Direct Test when 500 instead of 50 microliters of decontaminated sediment is used. J Clin Microbiol 34(1): 222-223.
- Bergmann JS, Yuoh G, Fish G, Woods G (1999) Clinical evaluation of the enhanced Gen-Probe Amplified Mycobacterium tuberculosis Direct Test for rapid diagnosis of tuberculosis in prison inmates. J Clin Microbiol 37(5): 1419-1425.
- Chedore P, Jamieson FB (1999) Routine use of the Gen-Probe MTD2 amplification test for detection of Mycobacterium tuberculosis in clinical specimens in a large public health mycobacteriology laboratory. Diagn Microbiol Infect Dis 35: 185-191.
- Chedore P, Broukhanski G, Shainhouse Z, Jamieson F (2006) Falsepositive amplified Mycobacterium tuberculosis Direct test results for samples containing Mycobacterium leprae. J Clin Microbiol 44: 612-613.
- Dowdy DW, Maters A, Parrish N, Beyrer C, Dorman SE (2003) Costeffectiveness analysis of the Gen-Probe Amplified Mycobacterium tuberculosis Direct test as used routinely on smear-positive respiratory specimens. J Clin Microbiol 41(3): 948-953.
- Barrett A, Magee JG, Freeman R (2002) An evaluation of the BD ProbeTec ET system for the direct detection of Mycobacterium tuberculosis in respiratory samples. J Med Microbiol 51: 895-898.
- Rüsch-Gerdes S, Richter E (2004) Clinical evaluation of the semiautomated BDProbeTec ET System for the detection of Mycobacterium tuberculosis in respiratory and nonrespiratory specimens. Diagn Microbiol Infect Dis 48: 265-270.
- Little MC, Andrews J, Moore R (1999) Strand displacement amplification and homogeneous real-time detection incorporated in a secondgeneration DNA probe system, BDProbe- TecET. Clin Chem 45: 777-784.
- McHugh TD, Pope CF, Ling CL, Patel S, Billington OJ, et al. (2004) Prospective evaluation of BD ProbeTec strand displacement amplification (SDA) system for diagnosis of tuberculosis in non-respiratory and respiratory samples. J Med Microbiol 53: 1215-1219.
- Bergmann JS, Keating WE, Woods GL (2000) Clinical evaluation of the BDProbeTec ET system for rapid detection of Mycobacterium tuberculosis. J Clin Microbiol 38: 863-865.
- Kievits T, Gemen BV, Strijp DV, Schukkink R, Dirks M, et al. (1991) NASBA isothermal enzymatic in vitro nucleic acid amplification optimized for the diagnosis of HIV-1 infection. J Virol Methods 35: 273-286.
- Ruiz P, Gutierrez J, Zerolo FJ, Casal M (2002) Genotype mycobacterium assay for identification of mycobacterial species isolated from human clinical samples by using liquid medium. J Clin Microbiol 40: 3076-3078.
- Viinanen AH, Soini H, Marjamaki M, Liippo K, Viljanen MK (2000) Ligase chain reaction assay is clinically useful in the discrimination of smearpositive pulmonary tuberculosis from atypical mycobacterioses. Ann Med 4: 279-283.
- Palacios JJ, Ferro J, Ruiz Palma N (1998) Comparision of the ligase chain reaction with solid and liquid culture media for routine detection of Mycobacterium tuberculosis in nonrespiratory specimens. Eur J Clin Microbiol Infect 17: 767-772.
- Tortoli E, Lavinia F, Simonetti MT (1997) Evaluation of a commercial ligase chain reaction kit (Abbott LCx) for direct detection of Mycobacterium tuberculosis in pulmonary and extrapulmonary specimens. J. Clin. Microbiol 35: 2424-2426.
- Laffler TG, Carrino JJ, Marshall RL (1993) The ligase chain reaction in DNA-based diagnosis. Ann Biol Clin 50: 821-826.
© 2018 Gülnur Tarhan. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






