- Submissions

Full Text
Approaches in Poultry, Dairy & Veterinary Sciences
Simultaneous Genetic Screening of Bovine Leukocyte Adhesion Deficiency, Complex Vertebral Malformation, Deficiency of Uridine Monophosphate Synthase, and Mule foot in Holstein Cows in Taiwan
Chun Hsuan Chao*, I Ming Chen, Yi Hsin Yeh, Kuo Hua Lee and Tsung Yi Lin
Livestock Research Institute, Council of Agriculture, Taiwan
*Corresponding author: Chun Hsuan Chao, Livestock Research Institute, Council of Agriculture, Taiwan
Submission: April 12, 2021;Published: April 23, 2021

ISSN: 2576-9162 Volume8 Issue3
Abstract
This study analyzed the prevalence of genetic defect carriers of Bovine Leukocyte Adhesion Deficiency [BLAD], Complex Vertebral Malformation [CVM], Deficiency of Uridine Monophosphate Synthase [DUMPS], and mule foot in the Holstein cow population in Taiwan. In total, 1688 cows from 31 herds were analyzed and 9 [0.53%] and 30 [1.78%] were BLAD and CVM carriers, respectively. No DUMPS or mule foot carriers were discovered. Frozen semen of genetic defect-carrying bulls is no longer imported to Taiwan in accordance with import restrictions on frozen bull semen implemented in 2005. However, pedigree analysis revealed that some defective alleles remain in Taiwanese dairy herds because of the mating program. The main source of the defect genotype is dam carriers. Close monitoring is thus necessary for eliminating the recessive genetic defects from the Holstein cattle population in Taiwan.
Keywords:BLAD; CVM; DUMPS; Mule foot; Holstein cattle
Introduction
Autosomal recessive defects in Holstein cows cause considerable economic losses in dairy farming. Thus, comprehensive cattle screening for genetic defects is essential. Conventionally, single-gene detection is used for such screening. However, simultaneous detection of all known genetic defects is essential for effectively preventing further transmission of defective alleles in the cattle population [1]. Zhang et al. [2] suggested that it is better to choose multiplexed chip-based technology for high-throughput genotyping of large numbers of genomic SNP [2]. The Igenity-Prime 50k SNP chip, produced by Neogen Corp. [Lincoln, NE, USA] and certified by the Council of Dairy Cattle Breeding [CDCB, USA], includes nearly 42000 genetic markers that enable highly accurate genomic testing. This chip simultaneously provides economically relevant breeding information as well as genotypic evaluation results for conditions such as BLAD, CVM, DUMPS, and mule foot. It’s a high throughput and uses a popular allele discrimination method to generate allele specific products [3,4]. We recently analyzed carrier frequencies of the genetic defect bovine brachyspina [BS] in the Holstein cows in Taiwan and corrected the contrasting case of brachyspina genotype [5].
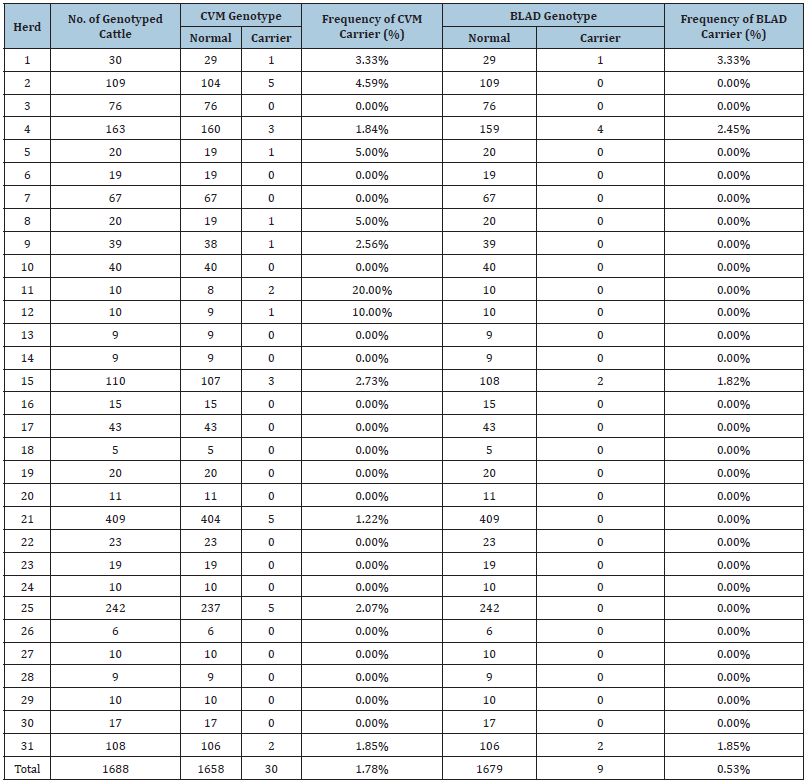
In Taiwan, no studies have focused on the prevalence of BLAD, CVM, or mule foot carriers among cattle, and only one reported the carrier frequency of DUMPS [6]. We determined the carrier frequencies of the aforementioned genetic defects in Holstein cows from several herds in Taiwan through genomic testing and investigated the transmission of these defects (Table 1).
Table 1:CVM and BLAD carrier frequencies among 31 herds of Holstein cows in Taiwan.
Materials and Methods
Hair follicle or blood samples were collected from 1688 random cows from 31 herds in sample collection cards and processed at 75 °C for 30 minutes. The animal use protocol listed above has been reviewed and approved by the Institutional Animal Care and Use Committee of Livestock Research Institute. The documents were accompanied by the sampled cows’ information including birthdate, sire, dam, and granddam. The sample collection cards were mailed to Neogene Corp., which tested the samples with Igenity-Prime 50k SNP chips. Sample processing and genotype analysis were performed in a CDCB-certified laboratory, and the data were emailed to the CDCB for genetic evaluation. The genetic evaluation results included key traits, production traits, reproductive traits, type traits, genetic defects, and parental confirmation. The genotype information for determining the sires of the genetic defect carriers was obtained from https://www. naab-css.org/dairy-cross-reference.
Result
We subjected samples from 1688 cows from 31 herds to genomic testing and simultaneously screened potential carriers for multiple genetic defects. No CVM carrier was detected in 19 herds. One to five carriers were noted in the other herds, with carrier frequencies of 1.22%-20%. The CVM carrier frequency was 1.78% [30/1688]. For the BLAD genotype frequencies, only four herds contained BLAD carriers, and one to four carriers were noted in these herds for carrier frequencies of 1.82%-3.33%. The BLAD carrier frequency was 0.53% [9/1688]. By contrast, no DUMPS or mule foot carriers were discovered in the current study.
Discussion
The simultaneous detection of multiple genetic defects may reduce testing costs and time and gain more amount of information from one experiment. The Ingenuity-Prime 50k SNP chip, comprising nearly 42,000 genetic markers, enables highly accurate testing for forecasting of economically relevant breeding information and genetic screening for conditions such as BLAD, CVM, DUMPS, and mule foot. The prevalence of BLAD, CVM, DUMPS, and mule foot alleles among cattle has been reported in many countries; however, only one such study, on the DUMPS allele [6], was conducted in Taiwan. Our goal is to prevent dairy cattle with genetic defects from breeding, thus reducing losses to the dairy farming industry.
CVM carrier frequency in cattle has also been reported in the decade in Brazil [1.53%][7], China [2.90%][8], Lithuania [1.00%] [9], Poland [16.62%][10], Macedonia [1.10%][11], Mexico [10.30%] [12]. BLAD carrier frequency in cattle has also been reported in the decade in Brazil [0.77%][7], China [0.49%][13], Lithuania [0.50%] [9], Macedonia [2.20%][11], Mexico [0.00%][12]. Although through the monitoring and moreover developed their breeding programs that could reduce the prevalence of genetic defect [14]. The CVM and BLAD defect alleles were still in the Holstein cow population in Taiwan. Some herds even have higher CVM or BLAD defect prevalence. On the other hand, no DUMPS or mulefoot carriers were discovered in the current study that is similar to findings in Turkey [15], Poland [16], India [17], and Iran [18]. However, Lin et al. [6] reported a DUMPS carrier frequency of 0.14% in Taiwan.
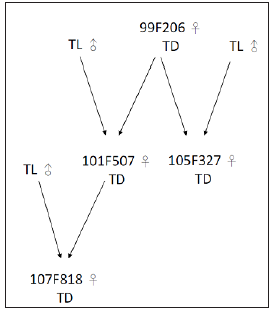
Our pedigree analysis demonstrated that the sire of all three CVM carriers, HOUSA000070625861, could be traced back to a common ancestor, the US Holstein sire Carlin-M Ivanhoe Bell (Table 2). Excluding 4 cows with unknown sires or sires without CVM genotype information, 88.46% [23/26] of carriers’ sires had the normal genotype for the CVM allele. In other words, the main source of the CVM genotype was the dams. Similar results were found for the BLAD genotype (Table 3). Excluding 1 cow with unknown sires, all sires [100%, 8/8] carried the normal genotype for the BLAD allele. In pedigree analysis for CVM and BLAD transmission, the defective allele was transmitted to three generations, despite the sires carrying the normal genotype. Such recessive allele transmission through dams was also noted in Poland [10]. Routine genetic defect screening of dairy cattle and the current restriction of frozen semen imports can effectively prevent the introduction of inferior livestock to Taiwan. Because the defective alleles no longer enter the dairy cattle population in Taiwan, we can infer that the main sources of the genetic defect are dam carriers. Although the carrier frequencies are <2%, close monitoring is crucial for eliminating defective alleles from Taiwanese dairy herds. Dairy farmers with herds containing high CVM or BLAD defect carrier frequencies should be counseled in appropriate mating strategies to gradually eradicate the defective alleles (Figure 1).
Table 2: Genotype of the sire of a CVM carrier.
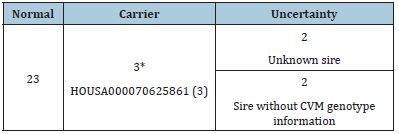
*From the Carlin-M Ivanhoe Bell family.
Table 3: Genotype of the sire of a BLAD carrier.
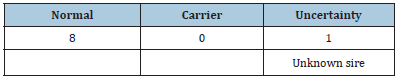
Figure 1:Pedigree analysis of BLAD genotype transmission. TL, BLAD-free animals; TD, BLAD carriers.
Acknowledgement
This work was financially supported by COA grants no: 108AS- 2.1.2-L1[1]. This manuscript was edited by Wallace Academic Editing.
References
- Kumar A, Gupta ID, Mohan G, Vineeth MR, Ravikumar D, et al. (2020) Development of PCR based assays for detection of lethal Holstein haplotype 1, 3 and 4 in Holstein friesian cattle. Molecular and Cellular Probes 50: 101503.
- Zhang Y, Liang D, Huang H, Yang Z, Wang Y, et al. (2020) Development and application of KASP assays for rapid screening of 8 genetic defects in Holstein cattle. Journal of Dairy Science 103(1): 619-624.
- Kim S, Misra A (2007) SNP genotyping: Technologies and biomedical applications. Annual Review of Biomedical Engineering 9: 289-320.
- Oliphant A, Barker DL, Stuelpnagel JR, Chee MS (2002) Bead array TM technology: Enabling an accurate, cost-effective approach to high-throughput genotyping. Biotechniques 32: S56-S61.
- Chao CH, Chen YM, Lee KH (2020) Genotype screening of bovine brachy spina in Taiwan Holstein cows. American Journal of Animal and Veterinary Sciences 15(3): 206-210.
- Lin DY, Huang YC, Chang HL, Liaw RB, Lee SC, et al. (2001) DNA typing of inherited deficiency of uridine monophosphate synthase in dairy cattle and beef cattle. J Chin Soc Anim Sci 30: 15-22.
- Paiva DS, Fonseca I, Pinto IS, Ianella P, Campos TA, et al. (2013) Incidence of bovine leukocyte adhesion deficiency, complex vertebral malformation, and deficiency of uridine-5-monophosphate synthase carriers in Brazilian Girolando cattle. Genet Mol Res 12(3): 3186-3192.
- Wang C, Tong Q, Hu XZ, Yang LG, Zhong XQ, et al. (2011) Identification of complex vertebral malformation carriers in Holstein cattle in south China. Genet Mol Res 10(4): 2443-2448.
- Morkūnienė K, Bižienė R, Pečiulaitienė N, Ugenskienė R (2019) Detection of bovine leukocyte adhesion deficiency, deficiency of uridine monophosphate synthase, and complex vertebral malformation in Holstein cattle. Slov Vet Res 56(2): 75-82.
- Ruść A, Hering D, Puckowska P, Barcewicz M, Kaminski S (2013) Screening of polish Holstein-Friesian bulls towards eradication of Complex Vertebral Malformation (CVM) carriers. Pol J Vet Sci 16(3): 579-581.
- Adamov N, Mitrov D, Esmerov I, Dovc P (2014) Detection of recessive mutations (BLAD and CVM) in Holstein-Friesian cattle population in Republic of Macedonia. Mac Vet Rev 37(1): 61-68.
- Virgen MA, Ayala VMA, Galindo GJ, Sánchez CDR, Lemus FC, et al. (2019) Carrier frequency of autosomal recessive disorders (BC, BLAD, FXID and CVM) in Holstein cows in Jalisco, Mexico. Pesquisa Veterinária Brasileira 39(7): 481-484.
- Li J, Wang H, Zhang Y, Hou M, Zhong J (2011) Identification of BLAD and citrullinemia carriers in Chinese Holstein cattle. Anim Sci Pap Rep 29: 37-42.
- Schütz E, Scharfenstein M, Brenig B (2008) Implication of complex vertebral malformation and bovine leukocyte adhesion deficiency DNA-based testing on disease frequency in the Holstein population. J Dairy Sci 91(12): 4854-4859.
- Meydan H, Yildiz MA, Agerholm JS (2010) Screening for bovine leukocyte adhesion deficiency, deficiency of uridine monophosphate synthase, complex vertebral malformation, bovine citrullinemia, and factor XI deficiency in Holstein cows reared in Turkey. Acta Vet Scand 52(1): 56.
- Kaminski S, Grzybowski G, Prusak B, Ruść A (2005) No incidence of DUMPS carriers in Polish dairy cattle. J Appl Genet 46(4): 395-397.
- Patel RK, Krishna MS, Kalpesh JS, Jenabhai BC, Krothapalli RS, et al. (2006) Lack of carriers of citrullinemia and DUMPS in Indian Holstein cattle. J Appl Genet 47(3): 239-242.
- Rahimi G, Nejati JA, Olek K (2006) Genotyping BLAD, DUMPS and κ-CSN loci in Holstein young bulls of the national animal breeding center of Iran. Pakistan J Biol Sci 9(7): 1389-1392.
© 2021 Chun Hsuan Chao. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






