- Submissions

Full Text
Approaches in Poultry, Dairy & Veterinary Sciences
The Mitochondrial Genome and Phylogenetic Relationships of Muong Lay Black Pig (Sus scrofa) in Vietnam
Thuy Thi Bich Vo1*, Tuan Anh Bui2, Hieu Duc Nguyen1, Hanh Hong Ha1 and Eui Bae Jeung3
1 Institute of Genome Research, Vietnam Academy of Science and Technology, Vietnam
2 Institute of Forensic Science, Vietnam
2 College of Veterinary Medicine, Chungbuk National University, Korea
*Corresponding author: Thuy Thi Bich Vo, Institute of Genome Research, Vietnam Academy of Science and Technology, 18, Hoang Quoc Viet, Cau Giay, Hanoi 100000, Vietnam
Submission: September 26, 2018;Published: October 11, 2018

ISSN: 2576-9162 Volume5 Issue1
Abstract
Muong Lay black pig is an indigenous breed of livestock and has recently been listed for conservation in Vietnam. The aim of this study was to investigate the phylogenetic relationship of the Muong Lay black pig with other pig breeds by sequencing its mitochondrial genome (mtDNA). The PCR products were sequenced by Applied Biosystems 3500 Genetic Analyzer. The complete mtDNA sequence was 16,742bp in length and includes two rRNA (12S and 16S), 22 tRNA, 13 mRNA genes, and one control region (D-loop). The phylogenetic analysis was performed by comparing both complete mtDNA and D-loop sequences of Asian and European pig breeds. The results of phylogeny reconstruction revealed that Muong Lay black pig belonged to an Asian clade and clustered with Bihu and Banmei pig from Yangtze River Region and Yellow River Region, while reporting distinctly different phylogenetic relationships with the European breeds.
Abbreviations: Mitochondrial DNA; Muong lay black pig; Pig breeds; Phylogenetic relationships; Sus scrofa
Introduction
Vietnam has numerous varieties of pig breeds distinct to each ecological region of the country. Muong Lay black pig is one of the local pig breeds with unique morphologic characteristics (e.g., dark back skin; straight back; 4-6 white spots on forehead, head, legs, and tail; and 14 teats) originally bred by the Muong community- a minority ethnic group residing in the mountainous region in Northern Vietnam. This is a voracious pig, well adapted to the harsh climate in term of high mountain zone, inadequate nutrition, and high disease resistance [1]. In addition, this pig breed provides excellent quality tenderloin and fragrant meat [2]. In recent years, the population size of Muong Lay black pig has been constantly decreasing [3] associated with high risks of losing rare allelic variations because of replacement by imported pig breeds. Consequently, in 2008, the Muong Lay black pig was included in the list of genetic resources that should be preserved in Vietnam [4]. The mitochondrial DNA (mtDNA) of vertebrates has become a common tool used in resolving phylogenetic relationships at different evolutionary depths levels related to particular properties, such as the presence of strictly orthologous genes, the lack of recombination, and an appropriate substitution rate [5]. For instance, mtDNA is deemed to strictly follow maternal inheritance and highly variable at the species level; the rate of mtDNA evolution is about 5 to 10 times higher than nuclear DNA, and hardly goes through gene recombination [6]. The fast rate of mutation produces more variance between sequences which gives an advantage when studying closely related species. Therefore, mtDNA sequences are useful markers for phylogenetic inference and also in studying genetic diversity [7]. The mitochondrial control region (Displacement or D-loop) is one of the fastest mutating sequence regions in animal DNA, and is also known to be more variable in sequence than other regions in either the nuclear or mitochondrial genomes [8]. There are several studies that have reported mutations in the D-loop sequence region for phylogenetic analyses [9-11]. For example, mtDNA from other Vietnamese pigs have been reported from I pig [12], Huong pig [13], Muong Khuong pig [14], and Mong Cai pig [15]. All of the previous studies reported the shortest genetic distance between the indigenous pigs of Vietnam and Asian pig breeds. The aim of this study was to derive the complete mtDNA sequence of the Muong Lay black pig breed to accurately assess the valuable genetic resources in Vietnam, and report on key implications for the maintenance and utilization of genetic diversity in Vietnamese livestock species. We constructed a phylogenetic tree of mtDNA data to investigate the evolutionary relationships between indigenous Muong Lay black pig and other wild boar and domestic pig breeds found in Asia and Europe.
Sample collection from all the Muong Lay black pigs used in this study was done in line with the guidelines of National Institute of Animal Sciences (Hanoi, Vietnam). Thirty blood samples (5ml per sample) were collected with anticoagulant from the jugular vein of Muong Lay black pigs reared in Muong Lay town, Dien Bien province (Vietnam). The experiments on animals were approved by the Ethics Committee at the Institute of Genome Research (Vietnam). Genomic DNA was extracted by standard phenol-chloroform method described by [16].
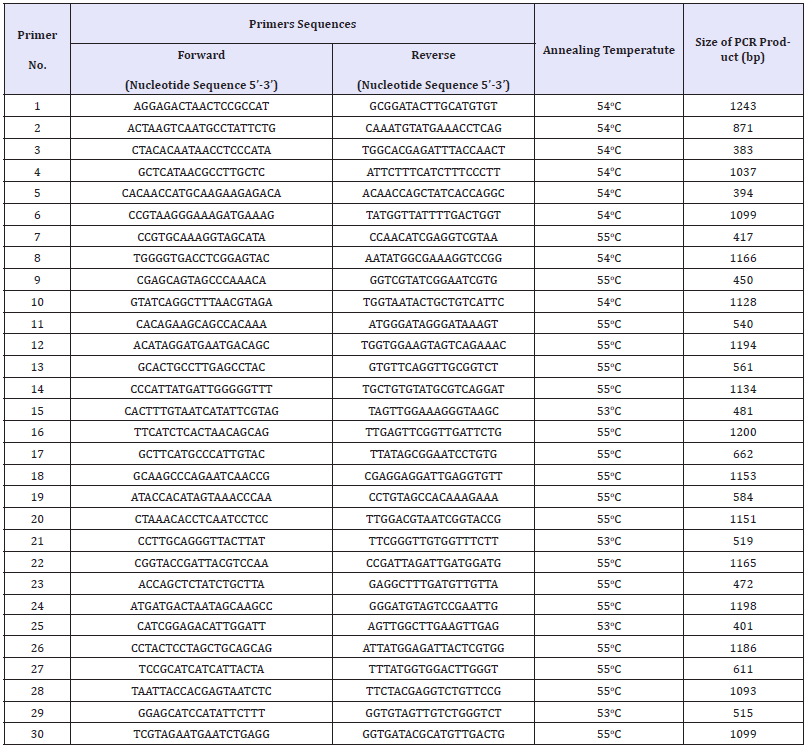
A total of 30 primer pairs (Table 1) was used to amplify the overlapping fragments of D-loop region and coding region sequences of pig mtDNA. The PCR reaction system was carried out in a 50μl volume, including 5μl of 10 x buffer (with Mg2+), 1μl of dNTP Mix, 1μl of 10mmol/L each primer, 0.5μl of 5U/μl Taq DNA polymerase and 2μl of genomic DNA. PCR-amplifications included an initial denaturation at 94 °C for 5 min, followed by 25 cycles, each one consisting of 30sec denaturation at 94 °C, 30sec primer annealing in range 53-55 °C (depending on the composition of primers), 30- 45sec extension at 72 °C, and then a final 8 min extension at 72 °C. Post amplified DNA was purified by GeneJET™ PCR Purification Kit (Thermo Fisher Scientific, Singapore, Singapore) and sequenced using ABI3500 Genetic Analyzer system (Applied Biosystems®, CA, USA).
Table 1:Primer pairs for Muong Lay black pig complete mitochondrial DNA.
The sequences were edited and assembled by DNADragon v1.6.0 software (SequentiX, Germany). The mitogenome sequence was annotated using BLAST, MITOS, and DOGMA web servers [17]. Base-composition statistics of the mitogenome were calculated using DAMBE v6.3.17 software [18].
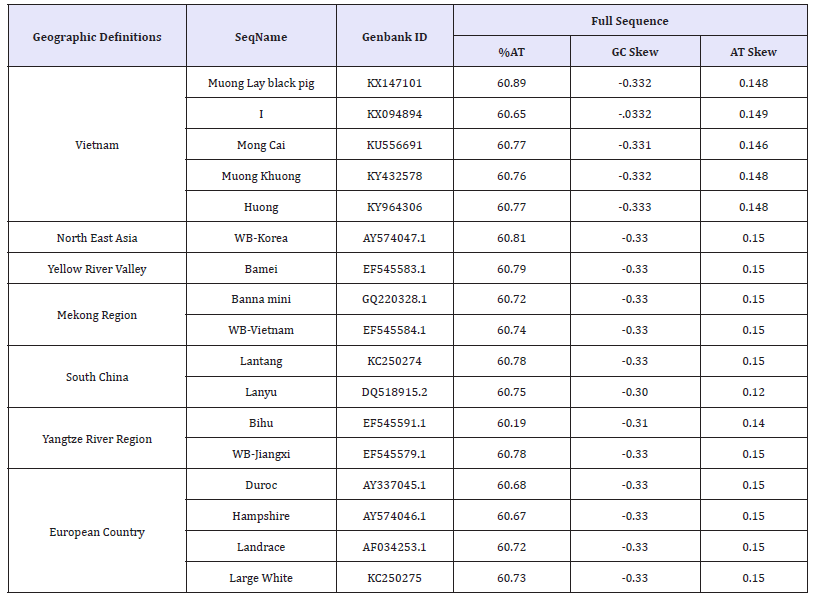
To analyse evolution of Muong Lay black pig breeds, 16 sequences were used, which were downloaded from the GenBank of the other domestic and wild board (WB) pigs in Vietnam and the rest of the world (Table 2). The mitochondrial sequence of African Warthog (Phacochoerus africanus)- a wild member of the pig family (Suidae) (accession number: DQ409327.1) was used as an outgroup. Multiple alignments of mtDNA sequences were performed using MUSCLE algorithm [19]. The best-fit models of nucleotide substitution was identified as TIM1+G for whole mtDNA sequence using jModelTest with NNI topology search in Cipres Gateway [20]. The alignment sequences were analysed in RAxML version 8.2.10 [21] to estimate the best maximum likelihood phylogenetic tree, with 1,000 bootstrap replicates, estimated GTRGAMMA model and the rapid bootstrapping algorithm. Finally, Fig. Tree v1.4.2 software was used to read exported format file for the phylogenetic tree construction.
Table 2:Pig breeds, their geographic definitions and sequence components.
Results and Discussion
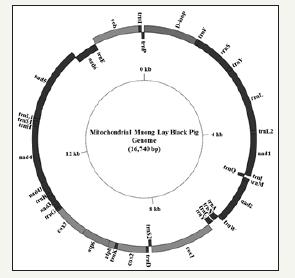
The complete mtDNA genome of Muong Lay black pig sequence from this study was deposited to the NCBI GenBank with accession number KX147101. The genome of Muong Lay black pig breed was 16,742bp in total length, containing 13 protein-coding genes, 22 transfer RNA (tRNA) genes, 2 ribosomal RNA (rRNA) genes and non-coding regions. The overall base composition was Adenosine (A) (34.90%), Cytidine (C) (26.09%), Guanosine (G) (13.02%), and Thymidine (T) (25.99%), and the percentage of A and T (60.89%) was higher than G and C percentage. The D loop region was 1,314 bp in length and located between tRNA-Phe and tRNA-Pro. The lengths of twenty two tRNA genes ranged from 59 to 75bp. The locations, size and anti codon of genes in the mtDNA genome are given in Table 1. Like other vertebrate animals, the order and composition of full mtDNA sequence of this indigenous Vietnamese pig were similar to most mitochondrial genomes [22-25]. Genes were encoded on the heavy strand except for ND6 and eight tRNA genes, which were on the light strand.
The nucleotide content of the entire mitogenome sequence of Muong Lay black pig was biased toward A + T rich that could be explained by the initiation of replication and transcription in accordance with mitogenomes of other Asia pigs [26]. The AT content was used to study the nucleotide compositional behaviour of mitochondrial genomes, and related to phylogenetic analysis [27], in order to show the difference of Muong Lay black pig component indexes, which may be linked to the little genetic variation of Muong Lay black pig as compared to the taxa of other pig breeds. In this study, the GC skew and AT skew were also calculated and used as indexes to estimate the genetic variant in finding the relatedness of phylogeny [28,29]. But the little difference of the GC skew and AT skew in Muong Lay black pig as compared to other pig breeds was not sufficient in drawing significant conclusion. There were seventeen repeat sequences (5’-tacacgtgcg) in the noncoding region that were different from other pig breeds such as Japanese Wild boar (1 repeat), Large White (6 repeats), Landrace (13 repeats) and Duroc (10 repeats). The larger the repeat motif number, the more chances for hairpin structures, leading to errors in the copying process [30].
Figure 1:The circular map of the mt genome of Muong Lay Black pig used GenomeVx.
As illustrated in phylogenetic trees of the complete coding sequence (Figure 1), the overview of the maximum likelihood topology showed that the bootstrap of nodes was higher than 70 bootstrap percentage (BP) except for the node of Lantang and Huong pigs (but it was still more than 50BP). Therefore the branches of the phylogenetic tree were well supported and the genetic relationships were certain [31]. The root of the phylogenetic tree was determined by using Warthog as an outgroup, which showed the independent development of two big groups of Sus scrofa depending on the geography, either in Asia or Europe [32,33]. In the Asian clade, Muong Lay and four other domestic pigs were joined to three subclades. In the first group, I pig was close to Banna mini pig and WB-Vietnam, which the bootstrap support of 100 and 91BP respectively. The second group contained Lantang pig and two Vietnamese pig breeds as Huong (bootstrap -54BP) and Mong Cai pig (bootstrap -97BP). The last group included Muong Lay, Muong Khuong pig (bootstrap -100BP) and two Chinese pig breeds are Bihu (bootstrap -83BP) and Bamei (bootstrap - 89BP). In addition, WB Jiangxi branch was between the first and second subclade (bootstrap - 97BP), while WB Korea and Lanyu pig fell out of three sub clades with bootstrap support of 98 and 95BP, respectively. These results showed the possibility that there may be other subclades.
Using the published sequence of pig breeds from different areas as reference points makes it easy to validate the results. Vietnam is an Asian country, so there is a high probability that Vietnamese domestic pigs were joined in the Asian clade. For instance, the I pig (“Ỉ” is the name of Vietnamese indigenous pig breed) had close genetic relationship with Banna mini - a pig breed from Mekong region in Vietnam. The Muong Lay black pigs and three other Vietnamese pig breeds were close to Chinese domestic pigs. While Huong and Mong Cai pig breeds were close to Lantang - a South China Region pig breed, other Muong Khuong and Muong Lay black pig breeds were close to Bihu and Bamei -breeds from two regions in Central China.
Based on the results shown in Figure 1, Muong Lay black pig breeds have close relationships with other pig breeds such as Muong Khuong (in Vietnam), and Bihu of Yangtze River Region (in China), with the bootstrap range of 83 to 100BP amongst these breeds. This study also found some interesting similarities in the morphological characteristics between the Muong Lay black pig and the pig breeds in the Yangtze River Region (e.g., small body weight around 50 to 60kg/individual mature; thin skin around 0.45cm; early maturing from 3 to 4 months old for female pigs or from 40 to 50 days old for male pigs; an average offspring size of 13 and seven pairs of teats) [34,35]. On the other hand, the European pig breeds (e.g., Landrace, Large-White, Hampshire, and Duroc) are usually over the 200kg body weight when mature, an offspring size of 10 to 12 individuals, and a maximum of six pairs of teats [36]. A hypothesis for these traits might be from thousands of years ago when pigs were traded by the nearby Vietnam-China border regions, where the Yangtze River Region pig breeds entered Vietnam and gradually developed into a Vietnamese domestic pig breed. However, it can be difficult to determine fully the original relationship between Muong Lay black pig, Bihu and pig breeds as genetic markers were not included in this study Figure 2 & Table 2,3.
Muong Lay black pig was also sister taxon with Bamei pig- a specific Yellow River region pig breed in East Asia region. These results may contribute additional evidence to the hypothesis about original East Asia of Vietnamese pig breeds [37]. In a previous study Hongo et al. [38] reported that Vietnamese pigs were genetically diverse and may be descendent of wild and domestic pigs from other regions of Asia. However, several wild boar subspecies inhabiting East Asia have been domesticated from WB, with such domestication occurring repeatedly from 6000 to 9000 years ago. Thus, our study suggest that Vietnamese pigs may share a common ancestor with other East Asian domestic pigs. However, further studies need to be carried out into genetic indicators such as synthetic biology, metabolic engineering, genome engineering, and genome editing [39] to affirm the origin of indigenous pigs in Vietnam.
Figure 2:The phylogenetic relationship was analysed Maximum likelihood inference in RAXML software version 8.2.10 and Fig. Tree version 1.4.2 by comparison of the mitogenome of Muong Lay black breed with other pig breeds. Abbreviations of pig breeds are the same in the Table 2.
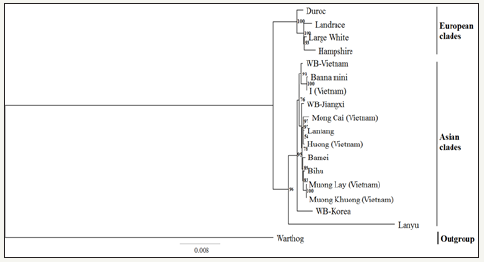
Table 3:Genes and their location in the mitochondrial genome of the Muong Lay black pig.
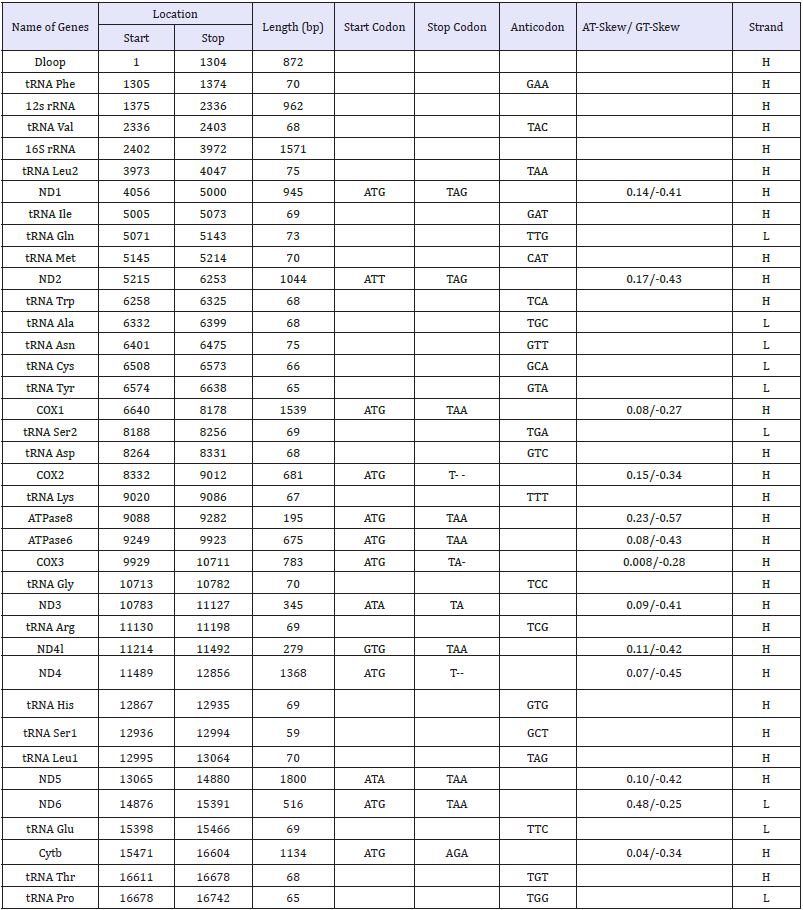
bp: Base Pairs; Heavy strand: H and Light strand: L; rRNA: Ribosomal RNA; rrnL: Large rRNA Subunit (16S); rrnS: Small rRNA Subunit (12S); tRNA: Transfer RNA and italic Words are Replaced by One Amino Acid Code; ND1-6 and ND4l: Genes Encoding Nicotinamide Dinucleotide Dehydrogenase Subunits 1 to 6 and 4l; ATPase6 and 8: Genes Encoding Adenosine Triphosphatase Subunits 6 and 8; COX1 to 3: Genes Encoding Cytochrome c Oxidase Subunits I to III; Cytb: Gene Encoding Cytochrome b. Intergenic Nucleotide Refers to the Nucleotide Distance Between Pairs of Adjacent Genes. Start and stop position of ribosomal RNA and non-coding region according to adjacent gene boundaries
TA- and T-- mean/ indicates the incomplete termination codon (the incomplete stop codon, that is, amino acid translation is terminated
when the gene forms a stop codon by post-transcriptional polyadenylation).
In the phylogenetic tree, Muong Lay black pig and other Vietnamese pig breeds also fell in the same sub-group as Chinese pigs, whereas it showed a significant difference to European pig groups. The Chinese domestic pig populations were derived from multiple Asian ancestral origins whereas the European domestic pig populations represent a single ancestral European lineage [40]. Therefore, Muong lay black pig probably belongs to the Asian type which differs from the European breeds.
Conclusion
By studying the mitogenome sequences and phylogenetic analysis of Muong Lay black pig in comparison to other Asian and European pig breeds, this study revealed a close relationship between the Muong Lay black pig breed and the pigs from Yellow River Valley region and Yangtze River Region in East Asia. A hypothesis was posed that the Muong Lay black pig’s origin belongs to East Asia region, but further research is needed for more clarity.
References
- Nguyen K (2009) Breeding the specialty pigs in the farm (in Vietnamese languge). Ministry of Agriculture and Rural Development, Hanoi, Vietnam.
- Trinh PN, Vo VS, Trinh PC (2011) Research and development of the local specialty pig breeds (Lung pigs and the 14 teat’s pigs of Muong Lay) in Phu Tho and Dien Bien Province (in Vietnamese language). National Institute of Animal Sciences, Hanoi, Vietnam.
- Vo VS (2004) Atlas of livestock breeds in Vietnam. Agricultural Publishing House, Hanoi, Vietnam, pp. 1-96.
- Dang Nguyen TQ, Tich NK, Nguyen BX, Ozawa M, Kikuchi K, et al. (2010) Introduction of various Vietnamese indigenous pig breeds and their conservation by using assisted reproductive techniques. J Reprod Dev 56(1): 31-35.
- Gissi C, Reyes A, Pesole G, Saccone C (2000) Lineage-specific evolutionary rate in mammalian mtDNA. Mol Biol Evol 17(7): 1022-1031.
- Castro JA, Picornell A, Ramon M (1998) Mitochondrial DNA: A tool for populational genetics studies. Int Microbiol 1(4): 327-332.
- Wolf C, Rentsch J, Hubner P (1999) PCR-RFLP analysis of mitochondrial DNA: a reliable method for species identification. J Agric Food Chem 47(4): 1350-1355.
- Nicholls TJ, Minczuk M (2014) In D-loop: 40 years of mitochondrial 7S DNA. Exp Gerontol 56: 175-181.
- Watanabe T, Hayashi Y, Ogasawara N, Tomoita T (1985) Polymorphism of mitochondrial DNA in pigs based on restriction endonuclease cleavage patterns. Biochemical Genetics 23(1-2): 105-113.
- Lan H, Shi L (1993) The origin and genetic differentiation of native breeds of pigs in southwest China: An approach from mitochondrial DNA polymorphism. Biochem Genet 31(1-2): 51-60.
- Gongora J, Fleming P, Spencer PB, Mason R, Garkavenko O, et al. (2004) Phylogenetic relationships of Australian and New Zealand feral pigs assessed by mitochondrial control region sequence and nuclear GPIP genotype. Mol Phylogenet Evol 33(2): 339-348.
- Nguyen HD, Bui TA, Nguyen PT, Kim OTP, Vo TTB (2017b) The complete mitochondrial genome sequence of the indigenous I pig (Sus scrofa) in Vietnam. Asian-Australas J Anim Sci 30(7): 930-937.
- Vo TTB, Nguyen HD, Bui TA, Nghiem MN, Jeung EB (2017) Phylogenomic analysis of mitochondrial DNA in the Huong pig: An indigenous pig of Vietnam. Global Journal of Animal Breeding and Genetics 5(4): 389-396.
- Nguyen DH (2017a) The Complete Sequence of Mitochondrial Genome of Muong Khuong Pig (Sus Scrofa). Int J Res Stud Biosci 5(7): 1-3.
- Tran TTN, Ni P, Chen J, Le TT, Steve K, et al. (2016) The complete mitochondrial genome of Mong Cai pig (Sus scrofa) in Vietnam. Mitochondrial DNA part B: Resources 1(1): 226-227.
- Sambrook J, Russell DW (2001) Molecular cloning: A laboratory manual. (3rd edn), Coldspring-Harbour Laboratory Press, UK .
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, et al. (2013) MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol 69(2): 313-319.
- Xia X, Xie Z (2001) DAMBE: Software package for data analysis in molecular biology and evolution. Journal of Heredity 92(4): 371-373.
- Edgar RC (2004) MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32(5): 1792-1797.
- Miller MA, Pfeiffer W, Schwartz T (2011) The CIPRES science gateway: A community resource for phylogenetic analyses. In TG ‘11 Proceedings of the 2011 TeraGrid Conference: Extreme Digital Discovery. ACM New York, NY, Salt Lake City, Utah, USA.
- Stamatakis A (2014) RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9): 1312- 1313.
- Yu G, Xiang H, Wang J, Zhao X (2013) The phylogenetic status of typical Chinese native pigs: Analyzed by Asian and European pig mitochondrial genome sequences. J Anim Sci Biotechnol 4(1): 9.
- Vo TTB, Nguyen HD, Bui TA, Nghiem MN, Jeung EB (2017) Phylogenomic analysis of mitochondrial DNA in the Huong pig: an indigenous pig of Vietnam. Global Journal of Animal Breeding and Genetics 5(4): 389-396
- Nguyen HD, Bui TA, Nguyen PT, Kim OTP, Vo TTB (2017) The complete mitochondrial genome sequence of the indigenous I pig (Sus scrofa) in Vietnam. Asian Australasian Journal of Animal Sciences 30(7): 930-937
- Nguyen HD (2017) Complete sequence of mitochondrial genome of Muong Khuong Pig (Sus Scrofa). International Journal of Research Studies in Biosciences 5(7): 1-3.
- Wang Y, Huang XL, Qiao GX (2013) Comparative analysis of mitochondrial genomes of five aphid species (Hemiptera: Aphididae) and phylogenetic implications. PLoS One 8(10): e77511.
- Song SN, Tang P, Wei SJ, Chen XX (2016) Comparative and phylogenetic analysis of the mitochondrial genomes in basal hymenopterans. Sci Rep 6: 20972.
- Hassanin A, Léger N, Deutsch J (2005) Evidence for multiple reversals of asymmetric mutational constraints during the evolution of the mitochondrial genome of Metazoa, and consequences for phylogenetic inferences. Syst Biol 54(2): 277-298.
- Wei SJ, Shi M, Chen XX, Sharkey MJ, van Achterberg C, et al. (2010) New views on strand asymmetry in insect mitochondrial genomes. PLoS One 5(9): e12708.
- Ghivizzani SC, Mackay SL, Madsen CS, Laipis PJ, Hauswirth WW (1993) Transcribed heteroplasmic repeated sequences in the porcine mitochondrial DNA D-loop region. J Mol Evol 37(1): 36-37.
- Soltis PS, Soltis DE (2003) Applying the bootstrap in phylogeny reconstruction. Statistical Science 18(2): 256-267.
- Fernández A, Alves E, Óvilo C, Rodríguez MC, Silió L (2011) Divergence time estimates of East Asian and European pigs based on multiple near complete mitochondrial DNA sequences. Anim Genet 42(1): 86-88.
- Giuffra E, Kijas JM, Amarger V, Carlborg O, Jeon JT, et al. (2000) The origin of the domestic pig: independent domestication and subsequent introgression. Genetics 154(4): 1785-1791.
- Nguyen DV (2012) Domestic pigs in Vietnam (in Vietnamese). Journal of Animal Science 11(23-26).
- Cheng P (1984) Livestock breeds of China. FAO, Rome, Italy.
- GCP/NEP/065/EC (2009) Farmer’s hand book on pig production. Food and Agriculture Organization of the United Nations, USA.
- Kim KI, Lee JH, Li K, Zhang YP, Lee SS, et al. (2002) Phylogenetic relationships of Asian and European pig breeds determined by mitochondrial DNA D‐loop sequence polymorphism. Anim Genet 33(1): 19-25.
- Hongo H, Ishiguro N, Watanobe T, Shigehara N, Anezaki T, et al. (2002) Variation in mitochondrial DNA of Vietnamese pigs: relationships with Asian domestic pigs and Ryukyu wild boars. Zoolog Sci 19(11): 1329- 1335.
- Paul O, Stephen H, Colin B (2014) Animal genetic resources. In: World Intellectual Property Organization (WIPO). FAO, Switzerland, pp. 1-178
- Luetkemeier ES, Sodhi M, Schook LB, Malhi RS (2010) Multiple Asian pig origins revealed through genomic analyses. Mol Phylogenet Evol 54(3): 680-686.
© 2018 Thuy Thi Bich Vo. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






