- Submissions

Full Text
Approaches in Poultry, Dairy & Veterinary Sciences
Investigation of Enterotoxigenic Escherichia coli Species Which Take Role in Calf Diarrhea and Detection of their Antimicrobial Susceptibility
KIRKAN Sukru*, PARIN Ugur, ASCI Ihsan Barıs and YUKSEL Hafize Tugba
Department of Microbiology, Adnan Menderes University, Turkey
*Corresponding author: PARIN Ugur, Adnan Menderes University, Faculty of Veterinary Medicine, Department of Microbiology, Aydin, TURKEY
Submission: April 25, 2018; Published: August 31, 2018

ISSN: 2576-9162 Volume5 Issue1
Abstract
The scope of this study was determination of enterotoxigenic Escherichia coli identification by detection of phenotypic and genotypic methods and determination of antibiotic susceptibilities of isolated strains from animals which had clinical findings such as diarrhea in Aydın province. Between September-December 2015, 50 calves with diarrhea symptoms were found in cattle farms in and around Aydın province. A total of 50 stool swabs taken from diarrheal calves were brought to the laboratory of the Department of Microbiology and Microbiology at the Adnan Menderes University Veterinary Faculty under the cold chain. Identification of was carried out by phenotypic methods from swab samples. The isolates identified as were genotypically confirmed by PCR and the presence of enterotoxigenic was also investigated by PCR. Antibiograms of strains identified in the last phase of the study were made by disk diffusion method. Isolation and identification of from 47 (94%) of 50 rectal swab samples examined. A total of 47 isolates (100%) examined after PCR using specific primers were found to be positive for uidA gene. Molecularly confirmed strains were identified as only 1 (2%) strains of enterotoxigenic when examined for the presence of ETEC-specific est1B gene. Antibiogram tests revealed that isolates were sensitive to 89% for Gentamycin, 61% for Cefoperazone, 51% for Amoxicillin-Clavulanic Acid, 44% for Danofloxacin, 42% for Enrofloxacin and Penicillin G and Erythromycin 100%, tetracycline and trimethoprim-sulfomethoxazole to 80% and Kanamycin to 76.5%. As a result, 94% of and 2% of these isolates were identified by phenotypic and genotypic methods from rectal swab samples taken from diarrheal calves in our study. In conclusion of antibiogram tests of isolates, multiple antibiotic resistance development was observed.
keywords:Enterotoxigenic ; Calf diarrhoea; Identification; PCR
Introduction
Bacteriologically-originated digestive system diseases widely seen all over the world are still an important health problem that affects both human health and animal health negatively. These diseases have an important cause of economic loss. Known as “Calf Diarrhea” or “Calf Septisemia” in the literature, colibasillosis is encountered in the calves during 2-10 days after birth. Escherichia coli is the causative agent of the disease that can be found in every enterprises. This bacteria is always abundant in the environment and in the intestines of healthy animals. The disease is more likely to occur in immunosuppressed animals due to inadequate care and nutritional conditions. The immunosuppressed calf intakes the bacteria through the respiratory, mouth, or umbilical cord. In sick calves, decrease in body temperature, yellow-whiteish and odorless watery diarrhea symptoms is seen in the tail parts of sick calves. However, there is a need for laboratory diagnosis in order not to interfere with diarrhea caused by other factors. For this purpose, if the calf is dead, it should be sent to the laboratory without being subjected to putrification. Some of sick calves recover within 5-10 days. Some of them suddenly become sick and die within 6 hours after clinical symptoms [1].
is known to be a Gram negative, rod-shaped, mostly moving, aerobic/facultative anaerobic breeding microorganism found in the normal intestinal flora of mammals and poultry. While non-pathogenic strains usually do not cause infection, pathogenic strains can cause serious infections. Enterotoxigenic strains cause gastroenteritis and travel diarrhea in humans and urogenital system infections, colibacillosis and colisepticemia in animals [2,3]. It is known that wild birds such as seagulls, as well as domestic animals such as cattle and sheep play an important role in infecting pathogenic strains to humans [4,5]. The agent is able to infect humans with direct contact with faeces of various domestic and wild animals or consumption of contaminant foods (uncooked meat and unpasteurized milk products). Since people in contact with diseased or transporter animals are at risk, it is very important to prescribe and treat pathogenic agents, especially in healthy animals [6-8].
Although is a microorganism that constitutes the healthy intestinal flora, infection can occur with an increased number of bacteria. In addition, oral intake of pathogenic with faeces and contaminated feed and water is among the important reasons for the formation of the infection. In general, the intestinal tract has two primary mechanisms that control the number of . The first factor is known as bacterial interference between other bacteria in the flora and , while the other factor is known as inadequate passive immunity. In the newborn calves, the intestinal tract is sterile. However, microorganisms in the vaginal and perianal flora of the mother can colonize the intestinal tract very quickly through the mouth and rectum. Coliform group agents, lactobacilli and obligatory anaerobic bacteria form this colonization within a few hours after birth. Gram positive, anaerobic lactobacilli are nonpathogenic and usually found in the small intestine, they decrease the pH due to the acid they produce and therefore cannot grow. On the other hand, colostrum and milk to be fed to the calf contain specific and nonspecific antibodies synthesized in the mother against . Thus, the gastrointestinal bacterial flora is kept in equilibrium with the immunoglobulin taken by colostrum and milk. It is reported that nonhygienic conditions, stress factors, climate changes, irregularities in nutrition increase of burden in the intestinal canal [6].
The scope of this study was to identify enterotoxigenic agent which is one of the most important agent of calf diarrhoea by phenotypic and genotypic methods from calves with diarrhoea. It is aimed to determine the most effective antimicrobial drugs as a result of antibiotic susceptibility tests and to keep the treatment costs which play an important role in calf breeding at a minimum level also.
Material and Methods
Specimen collection
This research was carried out between September-December 2015 by visiting cattle enterprises in and around Aydın province. The animal material of the study was composed of 50 calves presenting the symptoms of diarrhoea. Samples were collected by swabs animals’ rectum and delivered to the Routine Diagnostic Laboratory of the Department of Microbiology, Department of Veterinary Medicine, Adnan Menderes University in cold chain. These samples were investigated for the presence of enterotoxigenic . Adnan Menderes University Animal Experiments Local Ethics Committee (ADÜ-HADYEK) dated 14.08.2015 and numbered 64583101/2015/092 have not found any objection to the investigation.
isolation
It was aimed to obtain pure Escherichia coli from the laboratory samples. For this, swab samples were enriched in MTSB and incubated for 24 hours at 42 ˚C. Blood agar, EMB, MacConkey agar passages were performed to investigate the phenotypic characteristics of the strains that were observed at the end of 24 hours. After 24 hours of incubation, Gram staining method was applied to colonies. Identification of strains were determined by carbohydrate fermentation, indole, methyl red, oxidase negative, H2S, Voges Proskauer and citrate reactions [9,10].
Standard strains
ATCC® 25922 and ATCC® 35469 were obtained from the American Type Culture Collection (ATCC).
Primers
Primers for a putative toxin gene of (uidA-F: 5’ ATGCCAGTCCAGCGTTTTT GC 3’, uidA-R: 5’ AAAGTGTGGGTCAATAATCAGGAAGTG 3’) (1487 bp) and a plasmid encoding heat stabile toxin (est1b-F: 5’ TGTCTTTTTCACCTTTCGCTC 3’, est1b-R: 5’ CGGTACAAGCAGGATTACAACAC3’) (171bp) of enterotoxigenic [11] were ordered from Macrogen® (Republic of Korea).
DNA isolation
DNA extraction from isolates was performed using a DNA Extraction Kit (Fermentas®, Lithuania) according to the manufacturer’s specifications. The concentrations of DNA obtained using the kit were determined with a micro-volume spectrophotometer (ProNano PN-913, Maestrogen®, Taiwan).
PCR
PCR mixtures were prepared in a total volume of 25μl, containing a final concentration 1×Taq enzyme buffer solution, 100mM KCl, 25mM magnesium chloride (MgCl2), 10mM dNTPs, 0.2μM of each primer and 5U Taq DNA polymerase (Genet Bio® Exprime Taq DNA Polymerase, Republic of Korea) for identification of uidA and est1b gene-specific products in enterotoxigenic isolates [11].
The PCR products were loaded on a 2% Tris-EDTA buffer agarose electrophoresis gel stained with 3μl ethidium bromide. The bands were compared to a GeneRuler 100-bp DNA ladder (Fermentas ®, Canada) and a 1-kb DNA ladder (Fermentas®, Canada) to determine the size of amplicons.
Antibiotic susceptibility test
The disc diffusion method was applied using Mueller-Hinton Agar (Merck Millipore®, Germany) to determine the antibiotic susceptibilities of enterotoxigenic isolates. The antimicrobial agents used in the antibiotic susceptibility testing were penicillin G, tetracycline, cefoperazone, trimethoprim-sulfamethoxazole, erythromycin, amoxicillin-clavulanic acid, gentamicin, enrofloxacin, kanamycin, danofloxacin.
Results
Phenotypic identification and biochemical characterisation
After gram-staining, the gram-negative bacilli were subjected to biochemical identification. Lactose-positive pink colonies on Mac- Conkey agar, which had a metallic-green sheen on the EMB agar, were preliminary identified as . Subsequently, mannitol, glucose, indole, methyl red positive; oxidase, H2S, Voges Proskauer and citrate negative colonies were identified as . As a result, isolation and identification of from 47 (94%) of 50 rectal swab samples examined.
figure 1:E. coli uidA specific PCR results. M: 1500bp DNA Ladder; P: Positive Control (E. coli ATCC 25922); N: Negative Control; 1-12: uiaA gene Positive Samples
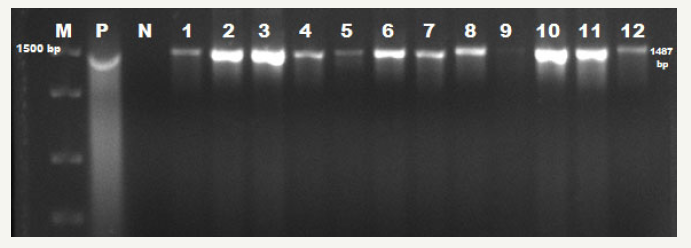
Genotypic identification
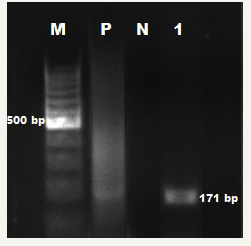
Forty-seven isolates identified as by biochemical identification profile exhibited expression of the uidA gene and were confirmed as . Only 1 enterotoxigenic was identified from strains examined after PCR performed using eST1B specific primers. Electrophoresis images obtained from the PCR are shown in Figure 1 & 2.
figure 2:ETEC est1B specific PCR results. M: 100bp DNA ladder; P: Positive Control (E. coli ATCC 25922); N: Negative Control; 1: ETEC est1B gene Positive Samples
Antibiotic susceptibility results
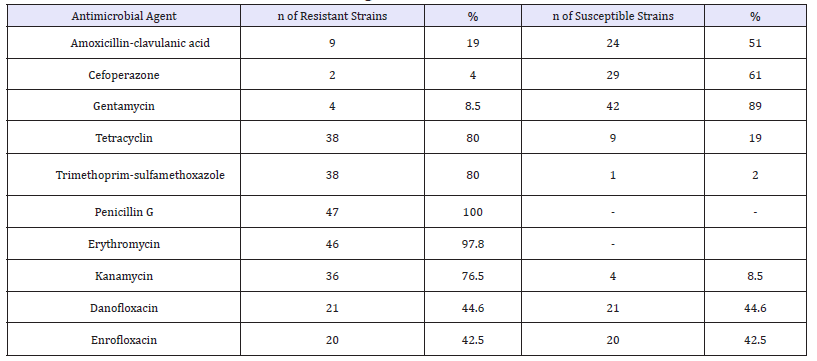
The antibiotic resistance rates determined against isolates are presented in Table 1. As a result of the antibiogram tests, the isolates of were susceptible to Gentamycin (89%), Sefoperazone (61%), Amoxycillin-Clavulanic acid (51%), Danofloxacin (44.6%), Enrofloxacin (42.5%) and resistant to Penicillin G (100%), Erythromycin (97.8%) Tetracycline (80%), Trimethoprim-Sulfomethoxazole (80%) and Kanamycin (76.5%).
Discussion
Enterotoxigenic strains can cause severe diarrhoea especially in young animals with enterotoxins [2]. Enterotoxigenic characteristics of strains isolated from enteric infections are determined by enzyme immunoassay (EIA), passive hemagglutination (PA), various latex agglutination tests, intestinal ligature testing in different animals, infantile mouse test (IFT), Vero, HeLa and Y1 cell cultures, rabbit vascular permeability test, DNA probing technique, radioimmunoassay (RIA), immunoblotting and different PCR techniques [12-14].
It has been reported that studies on the determination of enterotoxin types in strains isolated from different animals have been found in percentages between 1-66.7% of stabile toxin (ST) and 1.8-13.2% of labile toxin (LT) [8,14,15]. Carroll et al. (1990) reported that all of 16 ETECs from bovine origin (STIa) identified by DNA hybridization technique were found ST-positive BY competetive EIA and LT negative by latex agglutination test. In a similar study, Blanco et al. [15] reported that STa and LT were detected in one of the 197 strains isolated from diarrheal cows (STa) and LT in 2 (1.8%) in 3 isolates (2.7%) from healthy isolates. Rajkowa et al. [8] reported that 2 of the 54 strains originated from cattle (3.7%) were found to be ST gene positive and 1 (1.8%) strain was positive by PCR for LT gene. Mainil et al. [16] detected STb genes by PCR in 7 of 870 ETEC strains of cattle origin (0.8%). In another study, Shin et al. [17] reported that 7 of the 666 bovine isolates (1.1%) were found to be positive for the STb gene by DNA hybridization probe technique. In a study from Brazil, STA was detected in 8 (3.9%) and LT-II in 17 (8.3%) of 250 strains isolated from diarrheal calves [18]. In a similar study in the same country, Rigobelo et al. [19] showed that ST (25.4%) and LT (13.2%) genes were highly positive in calves with diarrhoea.
Table 1:The antibiotic resistance rates determined against E. coli isolates.
In a study conducted in Turkey, Güler et al. [14] found that 12 of 75 isolates from diarrheic calves of different farms were detected as enterotoxigenic and 16 of 45 isolates from healthy calves were negative by multiplex PCR. In a similar study, 390 strains isolated from healthy calves in Osek and Winiarczyk [20] were found to be negative by PCR in terms of LT and ST genes. Mills and Tietze (1984) identified stabile toxins by ELISA in 6 (2.4%) of 251 strains originating from calves. In another study Taku et al. [21] STa was detected in 31 (14.9%) calves with IFA, and STb was detected from 45 (21.6%) of 208 strains of calf origin by loop test. Wolk et al. [22] reported that 35 (58.3%) of 60 calf isolates were positive for ST by ELISA and 4 (66.7%) of 6 strains of lamb origin were positive for ST and all strains tested were negative for LT by the same method. In a study conducted in lambs, ST gene was determined by PCR in 2 (1.4%) of 146 strains [23].
The high prevalence of the geographic area and the incidence of species in calves were determined by Snodgrass et al. [24]. It was reported that season and geography play an effective role in the passive transfer of colostrum immunoglobulin in the calves [24]. It was reported that seasonal calf mortality was particularly important in immunoglobulin absorption in neonatal calves Fink [25]. One of the possible explanations for the high frequency of species during winter was the changes in barometric pressure along with climatic variables such as temperature, rain, and thunderstorms effective on the autonomic nervous system. Alternatively, another study suggests that high species in winter may be explained by the fact that the serum IgG1 concentration in winter is low and increases during spring and summer [26]. In our study, the transition of isolated strains from November to March appears to be seasonal. Herrera Luna et al. (2009) found that 17% of healthy and diarrheic calves were infected with in all of Austria. In addition, 15.2% of the strains carried the shiga toxin genes including the stx1, stx2, ehly and eae genes. There is another research which reported a low incidence of VTEC phenotype and O157: H7 serotypes of strains of diarrheal calves in Najaf, Iraq [27]. In one study, no cNF1, cNF2, stx2, stBand genes were found when the K99 fimbriae, stA enterotoxin, stx1 and eae genes were identified as 8, 8, 1 and 1 isolates, respectively [28]. Another study reported that the prevalence of other agents including Salmonella and Campylobacter species was detected 2% and 11% of the animals, respectively, while that of infected calves was in the ratio of 76%. In the same study, it was reported that 22/55 (40%) of diarrheal calves and 14/88 (16%) of strains from healthy calves carried K99 adhesin (P=0.001) [29]. In our study all isolates were identified as by uidA gene specific identification. Thus, it is seen that isolates from calf diarrhea are predominant bacterial species.
When the literature data are taken into consideration, it is reported that the rates of ST and LT positivity in strains among domestic animals are variable. In addition, studies in healthy animals have reported that these toxin types are found at low rates [8,15,17] and are generally negative [7,20]. In strains isolated from diarrheic animals, higher positivity is reported in various studies [18,19,30]. In this study where strains isolated from diarrheic animals were used, only 1 strain was detected as enterotoxigenic in 47 isolated from diarrheal specimens. Thus, strains are thought to originate from other serotypes.
Multidrug resistance in strains has been reported in previous studies [19,31,32]. Rigobelo et al. [19] showed that strains were the most resistant to cephalothin (46.1%), followed by Tetracy cline (45.7%), Trimetoprim-sulfadiazine (43.3%) and Ampisilin (% 41) were found to be resistant. Previous studies have shown that isolates have multiple resistances to beta-lactam antibiotics containing broad-range Aminoglycosides, Cephalosporins, Tetracycline, Sulfanomides and Fluoroquinolones [28]. De Verdier et al. [33], strains were found to be resistant to Sulfanomide, Streptomycin, Ampicillin and Tetracycline, while 61% of all strains were reported to be resistant to more than one antibiotic. Similar research has been conducted in Sweden De Verdier et al. [33], in the United States [34,35]. The prognosis for calf diarrhea due to septicaemia varies depending on how early the treatment is started and on the calf. In addition, although it is not a surprise that the selection of inappropriate antibiotics may increase the rate of resistance to antibiotics, more than 80% resistance to certain antibiotics has been detected in our research. Penicillin G and Erythromycin (100%), Tetracycline and Trimetoprim-Sulfomethoxazole 80%, and Kanamycin 76.5% resistance rates were detected in our study strains.
Conclusion
A total of 47 strains were identified as a result of isolation and identification studies of rectal swab samples collected from 50 diarrheal calves. Standard biochemical tests have shown that the isolates are molecularly confirmed by uidA, which is the control gene, and that it is fast and reliable in the identification of . Field strains that were confirmed as in the molecular context were subjected again to PCR testing for the presence of the ETEC est1b gene, resulting in only one enterotoxigenic identified. In addition, strains were found to have multiple antibiotic resistance and strains isolated from calf strains were found to have antibiotic resistance. It has been concluded that the high sensitivity to gentamycin is due to the fact that the antibiotic is not used on the ice due to its nephrotoxic effect. Amoxycillin-clavulanic acid combined preparations and cefoperazone (3rd generation) have been found as effective antibiotics against field strains.
Acknowledgement
This research was supported by the Adnan Menderes University Scientific Research Projects Committee (code No. VTF-15065).
References
- Radostits OM, Gay CC, Blood DC, Hinchcliff KW (1999) Veterinary medicine: A textbook of the disease of cattle, sheep, pigs, goats, and horses. (9th edn), WB Saunders, London.
- DebRoy C, Maddox CW (2001) Identification of virulence attributes of gastrointestinal Escherichia coli isolates of veterinary significance. Anim Health Res Rev 2(2): 129-140.
- Blanco M, Blanco JE, Dahbi G, Mora A, Alonso MP, et al. (2006) Typing of intimin (eae) genes from enteropathogenic Escherichia coli (EPEC) isolated from children with diarrhoea in Montevideo, Uruguay: identification of two novel intimin variants (muB and xiR/beta2B). J Med Microbiol 55(Pt 9): 1165-1174.
- Boynukara B, Solmaz H, Akgül Y, Aksakal A (2000) Yeni doğan buzağıların dışkılarında E. coli ve E. coli K99’un varlığı ile neonatal buzağı ishallerinin önlenmesinde oral Spektinomisin (Pentahidrat Dihidroklorit)’in etkisi. Bültendif 14: 2-5.
- Hossain MT, Siddique MP, Hossain FMA, Zinnah MA, Hossain MM, et al. (2008) Isolation, identification, toxin profile and antibiogram of Escherichia coli isolated from broilers and layers in Mymensingh district of Bangladesh. Bangl J Vet Med 6(1): 1-5.
- Çabalar M, Boynukara B, Gülhan T, Ekin IH (2001) Prevalence of Rotavirus, Escherichia coli K99 and O157: H7 in healthy dairy cattle herds in Van, Turkey. Turk J Vet Anim Sci 25: 191-196.
- Rajkhowa S, Hussain I, Rajkhowa C (2009) Detection of heat-stable and heat-labile enterotoxin genes of Escherichia coli in diarrheic fecal samples of mithun (Bos frontalis) calves by polymerase chain reaction. J Appl Microbiol 106(2): 455-458.
- Holt JG, Krieg NR, Sneath PHA, Staley JT, Williams ST (1994) Bergey’s Manual of Determinative Bacteriology, (9th edn), Williams & Wilkins, Baltimore, MS, USA, p. 179.
- Winn WC, Koneman EW, Allen SD, Jonda WM, Schreckenber PC (1997) Enterobacteriaceae, Color Atlas and Textbook of Diagnostic Microbiology, (5th edn), Lippincott Company, Philadelphia, New York, USA, pp. 171-241.
- Chandra M, Cheng P, Rondeau G, Porwollik S, McClellandb M (2013) A single step multiplex PCR for identification of six diarrheagenic E. coli pathotypes and Salmonella. Int J. Med. Microbiol 303(4): 210-216.
- Scotland SM, Flomen RH, Rowe B (1989) Evaluation of a reversed passive latex agglutination test for detection of Escherichia coli heat-labile toxin in culture supernatants. J Clin Microbiol 27(2): 339-340.
- Blanco JE, Blanco M, Mora A, Blanco J (1997) Production of toxins (enterotoxins, verotoxins and necrotoxins) and colicins by Escherichia coli strains isolated from septicemic and healthy chickens: Relationship with in vivo pathogenicity. J Clin Microbiol 35 (11): 2953-2957.
- Güler L, Gündüz K, Ok U (2008) Virulence factors and antimicrobial susceptibility of Escherichia coli isolated from calves in Turkey. Zoonoses Public Hlth 55(5): 249-257.
- Blanco M, Blanco J, Blanco JE, Ramos J (1993) Enterotoxigenic, verotoxigenic and necrotoxigenic Escherichia coli isolated from cattle in Spain. Am J Vet Res 54(9): 1446-1451.
- Mainil JG, Bex F, Jacquernin E, Pohl P, Couturier M, et al. (1990) Prevalence of four enterotoxin (STaP, STaH, STb and LT) and four adhesin subunit (K99, K88, 987P, and F41) genes among Escherichia coli isolates from cattle. Am J Vet Res 51(2): 187-190.
- Shin SJ, Chang YF, Timour M, Lauderdale TL, Lein DH (1994) Hybridization of clinical Escherichia coli isolates from calves and piglets in New York State with gene probes for enterotoxins (STaP, STb, LT), shiga-like toxins (SLT-1, SLT-11) and adhesion factors (K88, K99, F41, 987P). Vet Microbiol 38(3): 217-225.
- Salvadori MR, Valadares GF, Da Silva LD, Blanco J, Yano T (2003) Virulence factors of Escherichia coli isolated from calves with diarrhea in Brazil. Braz J Microbiol 34(3): 230-235.
- Rigobelo EC, Gamez HJ, Marin JM, Macedo C, Ambrosin JA, et al. (2006) Virulence factors of Escherichia coli isolated from diarrheic calves. Arq Bras Med Vet Zootec 58(3): 305-310.
- Osek J, Winiarczyk S (2001) Prevalence of eae and shiga toxin genes among Escherichia coli strains isolated from healthy calves. J Vet Med B Infect Dis Vet Public Health 48(1): 67-72.
- Taku A, Purohit VD, Sharma VK, Upadhyay SN (1991) Detection of K99 and F41 fimbria on ST enterotoxin producing Escherichia coli from calves. Indian J Anim Sci 61(3): 246-248.
- Wolk M, Ohad E, Shpak B, Adler H, Nahari N (1992) A survey of enterotoxigenic Escherichia coli from calves and lambs in the region of the Western Galilee in Israel during winter 1989-90. Israel Journal of Veterinary Medicine 47(1): 7-10.
- Orden JA, Ruiz Santa Quiteria JA, Cid D, Fuente RD (2002) Presence and enterotoxigenicity of F5 and F41 Escherichia coli strains isolated from diarrhoeic small ruminants in Spain. Small Rum Res 44(2): 159-161.
- Snodgrass DR, Terzolo HR, Sherwood D, Campell I, Menzies JD (1986) Aetiology of diarrhoea in young calves. Vet Rec 119(2): 31-34.
- Fink T (1980) Influence of type of housing, microclimate and management on health of calves. Hannover: Inaugural Disser Tierarztliche Hochshule.
- Norheim K, Simensen E, Gjestang KE (1985) The relationship between serum IgG levels and age, leg injuries, infections and weight gains in dairy calves. Nordisk Vet Med 37: 113-120.
- Al Charrakh A, Al Muhana A (2010) Prevalence of Verotoxin-producing Escherichia coli (VTEC) in a survey of dairy cattle in Najaf, Iraq. Iranian J Microbiol 2(3): 128-134.
- Bradford PA, Petersen PJ, Fingerman IM, White DG (1999) Characterization of expanded-spectrum cephalosporin resistance in E. coli isolates associated with bovine calf diarrhoeal disease. J Antimicrob Chemother 44(5): 607-610.
- Achá SJ, Kühn I, Jonsson P, Mbazima G, Katouli M, et al. (2004) Studies on calf diarrhoea in Mozambique: prevalence of bacterial pathogens. Acta Vet Scand 45(1-2): 27-36.
- Hammermueller J, Kruth S, Prescott J, Gyles C (1995) Detection of toxin genes in Escherichia coli isolated from normal dogs and dogs with diarrhea. Can J Vet Res 59(4): 265-270.
- Khan A, Das SC, Ramamurthy T, Sikdar A, Khanam J, et al. (2002) Antibiotic resistance, virulence gene, and molecular profiles of Shiga toxin-producing Escherichia coli isolates from diverse sources in Calcutta, India. J Clin Microbiol 40(6): 2009-2015.
- Dolejská M, Senk D, Cízek A, Rybaríková J, Sychra O, et al. (2008) Antimicrobial resistant Escherichia coli isolates in cattle and house sparrows on two Czech dairy farms. Res Vet Sci 85(3): 491-494.
- De Verdier K, Nyman A, Greko C, Bengtsson B (2012) Antimicrobial resistance and virulence factors in Escherichia coli from Swedish dairy calves. Acta Vet Scand 54: 2.
- Berge AC, Hancock DD, Sischo WM, Besser TE (2010) Geographic, farm, and animal factors associated with multiple antimicrobial resistance in fecal Escherichia coli isolates from cattle in the western United States. J Am Vet Med Assoc 236(12): 1338-1344.
- CLSI (Clinical and Laboratory Standards Institute) (2006) Performance Standards for Antimicrobial Disk Susceptibility Tests, Approved Standard-Ninth Edition (M2-A9). Clinical and Laboratory Standards Institute, Wayne, PA, USA.
© 2018 PARIN Ugur. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






