- Submissions

Full Text
Approaches in Poultry, Dairy & Veterinary Sciences
Physicochemical Properties of PI3K Inhibitor and Its Relation with the Structure and Activity
Gonzalez MTP1* and Wester JVWC2
1 School of Pharmaceutical Sciences of Ribeirão Preto, University of São Paulo, Brazil
2 Department of Genetics, Ribeirão Preto Medical School, University of São Paulo, Brazil
*Corresponding author: Gonzalez MTP, School of Pharmaceutical Sciences of Ribeirão Preto, University of São Paulo, Ribeirão Preto-SP, Brazil
Submission: March 15, 2018; Published: August 03, 2018

ISSN: 2576-9162 Volume4 Issue4
Abstract
The physicochemical properties of the fifty-seven molecules consider as inhibitors of PI3K were calculated and evaluated by descriptive statistics and principal component (PCA) analyses. PCA reduced physicochemical variables to two independent components. Here we show that six common characteristics (HBD, Dbonds, LogP, MR, S-bonds and TPSA) have low variability and could be the important characteristics that describe this type of molecules. In addition, the structural similarities as the presence of pyrimidine and triazine rings may be related to compound activity on cancer proteins like PI3K. These physicochemical and structural properties could be taken into account for anti-cancer treatments.
keywords: PI3K; Principal component analysis; Variables
Introduction
The PI3K protein is a phosphoinositide 3-kinase that plays a central role in the PI3K-AKT-mTOR signaling network responsible for cell proliferation control. Several mutations in proteins related to cell proliferation can deliver in tumor proliferation by triggering cancer-signaling pathways. In human cancer, cells which presents modifications in the PI3K protein structure result in a wrong production of the enzymes subunits α, β, γ and δ, which are the most common and important for human cancer development. Since the activation of the catalytic site of these subunits can occurs from a vastly number of stimuli there is a uniformity on the mechanisms that can triggers metastasis [1]. In order to stop cancer progression, the inhibition of PI3K proteins has been studied extensively using organic and inorganic compounds that can directly interact to the protein to stop the uncontrolled proliferating state of the cell. The first step
to find a possible inhibitor is the search of compounds with physicochemical and structural characteristics that permits to bind this protein [2]. In this context, Principal Component Analysis (PCA) is a useful tool for exploratory data analysis for example to study the relationship between the molecular activity and physicochemical properties of possible PI3K inhibitors. Our aim was to characterize the physicochemical properties of 57 PI3K inhibitors of which, [3- 5] and identify if these physicochemical propierties together with the chemical structure are associated to biological activity Figure 1.
We obtained the physicochemical properties by using the package ChemMine Tools, and a PCA was applied after standard ization of physicochemical properties of fifty-seven molecules. We obtained twelve chemical variables placed into two principal components with eingen values greater than 1.0, which is a common statistical cut-off point [6]. The two principal components accounted for 60.76% of the total variance. Thus, the dimensionality of the data was reduced from ten variables to two uncorrelated principal components with a 15.78% loss of variation.
Dim1 was high occupied by HBA (Hydrogen binding aceptor), HBD (Hydrogen binding donor), atoms and MR (Molar refrativity) while Dim 2 had the greatest values for TPSA (total polar surface area) and HBD (Hydrogen binding donor, that has the highest correlation) Figure 2. Fifteen molecules (1, 2, 6, 10, 12, 15, 18, 20, 21, 22, 26, 27 35, 36, 37) are in a decreasing classification in relation to the values from the most correlated variables (HBD, Dbonds, LogP, MR, S-bonds and TPSA). These molecules have in common the presence of linear chains or several halogens in their structure and almost all physicochemical properties have a state always below the general average that resulted in negative values for Dim 1 and 2. Only molecule named 31 has an increasing classification in relation to the values from the variables and all physicochemical properties values are over general overage.
At the structural level was observed that molecules with positive values on dim 1 and 2 have in common the presence of heterocyclic rings (specifically pyrimidine and triazine ring), which in addition are binding to high electronegative element and according to Peng et al. [3] due to in the majority of these molecules is the interaction site to the adenine pocket of kinase protein.
Conclusion
In conclusion, six variables (HBD, Dbonds, LogP, MR, Sbonds and TPSA) (Figure 1), have the best correlation due to values with the least variation. These observations could represent important values for molecules against PI3K activity. Additionally, heterocyclic rings like pyrimidine or triazine interact with the active site in an adenine pocket of kinase protein. Further analysis must be performed with an increased number of molecules, which could help to standardize a protocol to the discovery of novel PI3K activity inhibitors and to contribute for future anti-cancer therapies.
Figure 1: PCA graphic of molecules with PI3K inhibition activity. On the left are plotted molecules distribution and on the right are plotted the variables.
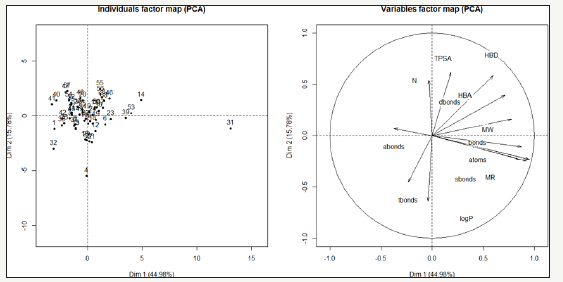
Figure 2: Dispersion graphics of variables values.

References
- Fruman DA, Rommel C (2014) PI3K and cancer: lessons, challenges and opportunities. Nat Rev Drug Discov 13(2): 140-156.
- Thorpe LM, Yuzugullu H, Zhao JJ (2015) PI3K in cancer: divergent roles of isoforms, modes of activation and therapeutic targeting. Nature Reviews Cancer 15(1): 7-24.
- Wu P, Nielsen TE, Clausen MH (2015) FDA-approved small-molecule kinase inhibitors. Trends Pharmacol Sci 35(7): 422-439.
- Cascioferro S, Parrino B, Spanò V, Carbone A, Montalbano A, et al. (2017) 1,3,5-Triazines: A promising scaffold for anticancer drugs development. Eur J Med Chem 142: 523-549.
- Sun T (2015) A Systematic Review on Antitumor Agents with 1, 3, 5-triazines. In: Zhixu Z, Du L (Eds.), Medicinal Chemistry JOUR, OMICS International, USA.
- Backman TW, Cao Y, Girke T (2011) ChemMine tools: an online service for analyzing and clustering small molecules. Nucleic Acids Res 39: 486- 491.
© 2018 Hantanirina Herisoa Isabelle. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






