- Submissions

Full Text
Approaches in Poultry, Dairy & Veterinary Sciences
Identification of the Staphylococcus Species Which Cause Cattle Mastitis Using Maldi-TOF MS
Kırkan Sukru1*, Parın Ugur1, Tanır Tansu2 and Yuksel Hafize Tugba1
1Department of Microbiology,Adnan Menderes University,Turkey
2Department of Microbiology,Adnan Menderes University,Turkey
*Corresponding author: Kırkan Sukru,Department of Microbiology,Faculty of Veterinary Medicine,Adnan Menderes University,Aydin, Turkey
Submission: May 21, 2018;Published: July 11, 2018

ISSN 2576-9162Volume4 Issue2
Abstract
The aim of this study was identification and antibiotic susceptibility of Staphylococcusspecies from mastitic milk, comparison of results by using commercial identification methods such as VITEK, MALDI-TOF MS analysis. A total of 100 milk samples taken in sterile conditions were investigated for the presence of Staphylococcus sp., which were found to have clinical mastitis problems in animals in the milk province of Aydın province and surrounding areas. Identification of Staphylococcus species was performed by VITEK 2, VITEK (MALDI-TOF) MS in addition to biochemical methods. Staphylococcus sp. was isolated from 34% of the milk samples. Seven (20.60%) of the isolates were Staphylococcus aureus, 10 (29.40%) were Staphylococcussimulans, 8 (23.50%) were Staphylococcus epidermidis and 4 (11.80%) Staphylococcus saprophyticus, 2 (5.90%) were Staphylococcuschromogenes, 2 (5.90%) were Staphylococcus hyicusand 1 (2.95%) were Staphylococcus arlettae. result of the antibiogram test, 34 Staphylococcus sp. isolates were sensitive to Cefoxitin, Ciprofloxacin, Vancomycin, Tigecycline and Fosfomycin in the ratio of 100% and resistant to Linezolide, Clindamycin and Tetracycline in the ratio of 85.2%, 100% and 82.4% respectively. In conclusion; the reliability of the MALDI-TOF MS proteomic biomass spectrophotometry method has been demonstrated to be verifiable and feasible.
Keywords: Mastitis; S aureus; Coagulase negative Staphylococcus; MALDI-TOF MS
Introduction
Mastitis is an important disease in terms of its prevalence and the economic losses it incurs due to the drop-in milk yield in the business. For this reason, it is important to prevent any damage that may be caused by early diagnosis of clinical mastitis, as in all diseases. Staphylococcal agents are the major bacterial agents causing bovine mastitis. These factors are spread widely, especially with staff and milking units. Staphylococci are predominant pathogens in mastitis cases of subclinical and clinical type. Staphylococci are able to adapt very rapidly to changes in environmental, host, mastitis control and care. Staphylococcal clinical cases are characterized by prolonged but relatively infrequent occurrence. Staphylococci are characterized by various pathogenic factors.
These factors are those that trigger damage to mammary tissues and neutralize antimicrobials that can escape immunity defences [1-3]. The virulence factors in the Staphylococci have been extensively researched. Staphylococcus aureus strains isolated from bovine mastitis cases include leukocidin, enterotoxin and coagulase in addition to alpha, beta, gamma, delta toxins. CNS strains also produce some toxins and enzymes (hemolysin, leukocidin, lipase, protease, DNase) that can be effective in disease formation. A large number of Coagulase Negative Staphylococci (CNS) isolated from mastitis cases show protease, DNase and lecithinase activity in excess, unlike the CNS isolated from normal cows. The isolation rate of S. aureus in previous studies performed in our country was 28.3% in Aydın, 40.1% in Afyon and 43% in Burdur [4-6].
Mastitis is an important mammary gland disease caused by microorganisms. The economic losses associated with mastitis are not only limited to a reduction in milk yield, but also include the treatment of the disease and the removal of diseased animals. In addition, deterioration in the quality of the milk, which is an important food source, causes negative results in terms of public health [4]. By means of MALDI-TOF MS (Matrix Assisted Laser Desorption/Ionization with Time-of-Flight Mass Spectrometry) method, a specific kind of fingerprints for microorganisms based on mass spectrometry are obtained from proteins specific for each organism and bacterial, fungal and virus identification can be performed [7]. The matrix is necessary for successful ionization of the material as it acts both as an environment in which ionization can occur and as a proton supplier for the ionization of the material.
The sample-matrix crystal on the surface of the metal plate is irradiated using a UV laser beam. Irradiation occurs briefly to avoid degradation, which may be caused by overheating [8]. Bacterial identification is the result of comparing the information obtained from the sample with the information in the database [9]. It has been reported that the correct identification rate of MALDI-TOF MS in species-based routine bacterial isolation varies between 84.1% and 95.2% [10,11]. The scope of this study was to determine isolation and antibiotic susceptibility of the most common Staphylococcus species in mastitis cases which are very important for milk production, to biotype and isolate the isolated strains by commercial identification methods such as VITEK and MALDI TOF MS.
Materials and Methods
Sample collection
The study was carried out between March and August 2017 in dairy cattle breeding farms in private enterprises in Aydin province. One hundred milk samples from Holstein dairy cows which showed signs of clinical and subclinical mastitis in 8 farms with 25-250 head animal capacity were used. The animals chosen for sampling were in the lactation period, at 2-8 years of age, did not receive antibiotic treatment in one month of period. Adnan Menderes University Animal Experiments Local Ethics Committee was informed by ethics decision no. 64583101/2016/140 dated 25.08.2016 VII session.
Staphylococcus sp. isolation
Milk samples brought to the laboratory were directly inoculated in Mannitol Salt Agar (MSA). After 24 hours of incubation at 37 °C, the colonies were cultured in a blood agar containing 5% sheep blood. After 24 hours incubation, the biochemical characteristics of the strains evaluated as Gram positive cocci by Gram staining were evaluated by standard laboratory procedures. Round shaped bacteria with Gram (+) character were searched on microscope [12]. Identification of Staphylococcus sp. isolates was carried out by catalase, DNase and coagulase reactions, mannitol fermentation and bacitracin susceptibility assays.
Identification of Staphylococcus sp. with VITEK 2 compact®
In this study, VITEK GP (Gram Positive) identification cards were which included Staphylococci in test database. From 18-24h fresh cultures grown in 5% sheep blood agar, the suspension was prepared with sterile cotton swab sticks and with a density of 0.50-0.63 McFarland in 3ml sterile saline solution according to the manufacturer’s instructions. In the McFarland measurement, the bioMérieux® brand DensiCHEK™ Plus model McFarland was used. The prepared bacterial suspension tubes were loaded with VITEK GP® cards and loaded onto the filler of the device by matching the card barcodes and sample names in accordance with the manufacturer’s instructions. At the end of the filling process in the filler section, the cards were transferred to the incubation module of the device. After this transfer, the device again checks the entered sample names and card barcode’s mappings and the filled cards are automatically transferred to the incubation module of the device. At the end of the incubation period the results were automatically printed by VITEK 2 Compact®.
Identification of Staphylococcus sp. with VITEK (MALDITOF) MS
In this study VITEK MS/IVD/V.3.0 (bioMérieux, France) database was used and all the operations performed in the direction of the manufacturer were carried out as follows. Bacterium agar subculture was obtained from our staphylococcal isolates determined as Gram positive cocci and catalase positive by preliminary tests. One colony from fresh colonies after 18-24 hours was taken with a 1μL aliquot and spread as a thin layer on VITEK MS slides. On the same slide, 34 isolates were prepared. The calibration curve, located in the middle of each 16 wells on the prepared slide, was spread in a thin layer of the Escherichia coli ATCC 8739 with mass spectrum profile for quality control and calibration purposes. A 1μl matrix solution of CHCA (α-cyano-4-hydroxycinnamic acid) was added to each of the prepared wells and allowed to dry in the chamber. After waiting for 2 minutes, the slide was inserted into the device and the operation started. After the vacuum and calibration process lasted about 10 minutes, the spectra of each of the isolates began to be taken when the device was first turned on. The total reporting time of our slide with 48 samples and 3 calibration points was measured as 57 minutes (about 1.7 Minutes per sample).
Antibiotic susceptibility test
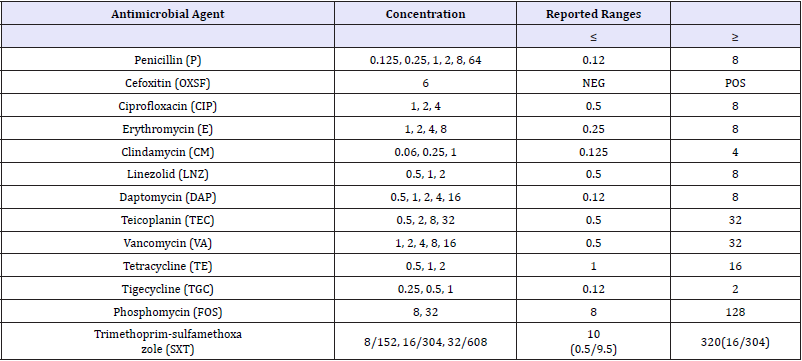
VITEK AST-P641 cards with reference number 418591 were used for antibiogram. The antibiotics, concentrations and reported ranges of VITEK AST-P641 are given in Table 1. A total of 280μL bacterial suspension at 0.50-0.63 McFarland concentration prepared in the VITEK GP cards was transferred to another sterile 3mL saline solution and mixed homogenously by automatic pipetting. As it is in the process of identification with VITEK Cards, it was transferred to the incubator module after it was filled by the device first. Results were received by the device within 8 hours.
Table 1: The antibiotics, concentrations and reported ranges of VITEK AST-P641
Results
Isolation and identification
Of the 100 milk samples examined in our study, 34 (34.0%) of the samples were isolated as Staphylococcus sp. As a result of coagulase test, 7 (20.6%) isolates were Coagulase Positive Staphylococci and 27 (79.4%) were Coagulase Negative Staphylococci respectively. Seven (20.6%) isolates were identified as Staphylococcus aureus, 11 (32,3%) isolates as Staphylococcus simulans in, 8 (23.5%) isolates as Staphylococcus epidermidis, 4 (11.8%) isolates as Staphylococcus saprophyticus, 2 (5.9%) isolates as Staphylococcus chromogenes, and 2 isolates (5.90%) as Staphylococcus hyicus in conclusion of biochemical tests.
Table 2: The results of conventional methods, VITEK 2 Compact and MALDI-TOF MS identification
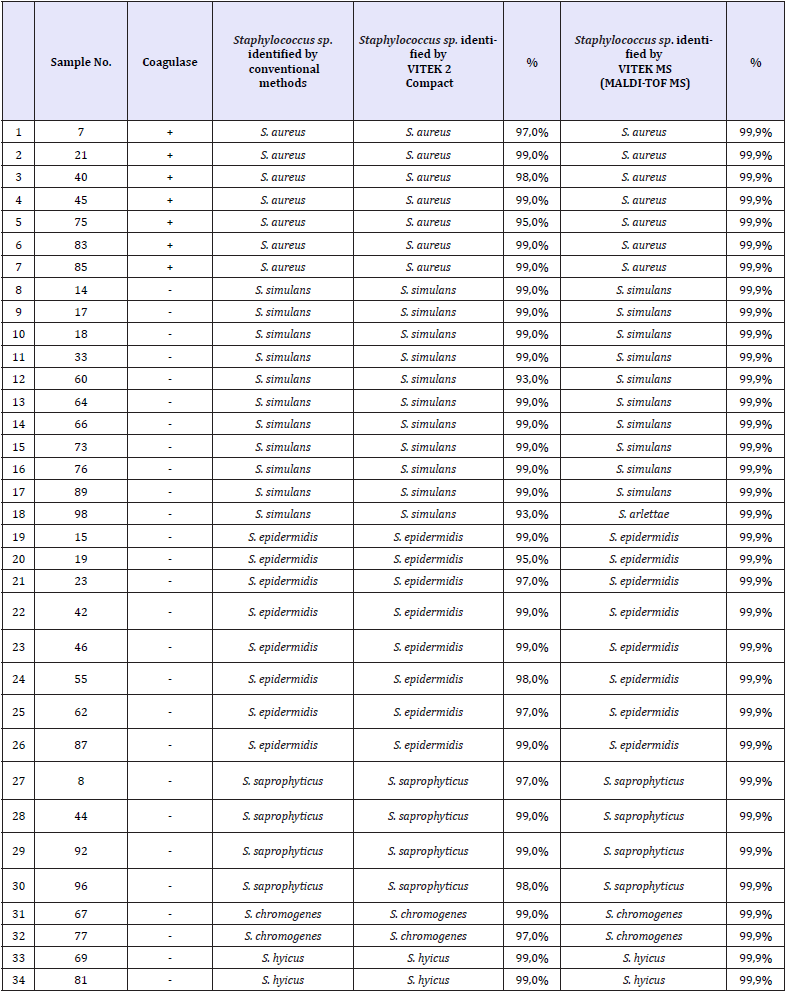
As a result of identification with VITEK 2 Compact, 7 (20.6%) Staphylococcus aureus, 11 (32.3%) Staphylococcus simulans, 8 (23.5%) Staphylococcus epidermidis, 4 (11.8%) Staphylococcus saprophyticus, 2 (5.9%) Staphylococcus chromogenes, 2 (5.9%) Staphylococcus hyicus were identified. As a result of identification studies with MALDI-TOF MS device, 7 (20,60%) Staphylococcus aureus, 10(29,40%) Staphylococcus simulans, 8 (23,50%) Staphylococcus epidermidis, 4 (1,80%) Staphylococcus saprophyticus, 2 (5,90%) Staphylococcus chromogenes, 2 (5.90%) Staphylococcus hyicus and 1 (2.95%) Staphylococcus arlettae were identified. The results obtained with the VITEK 2 Compact in our study are in agreement with the results obtained with conventional methods. When the results of MALDI-TOF MS are examined, it is seen that 1 Staphylococcus simulans isolate is identified as Staphylococcus arlettae (Table 2).
Antibiogram
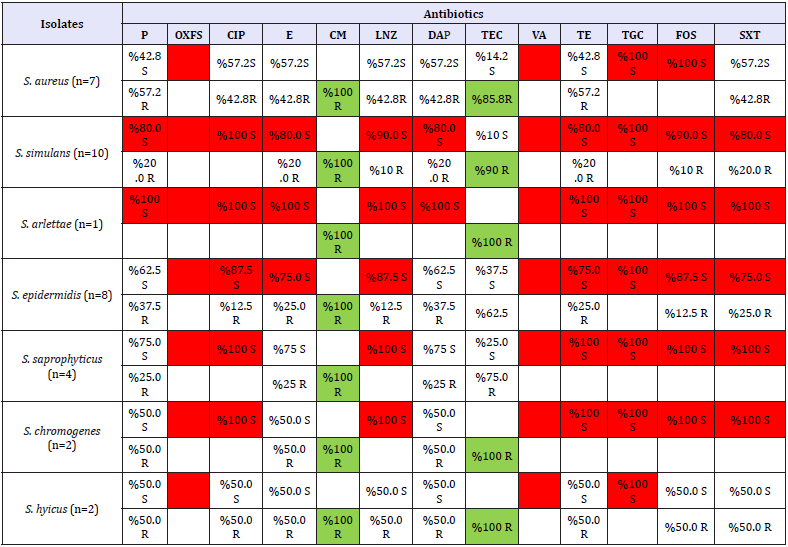
Staphylococcus strains which have been typed in our study were treated with VITEK AST-P641 reference no. 418591 and VITEK 2 Compact antibiogram device. The results of antibiograms evaluated according to CLSI (2017) standards of isolates are shown in Table 3 & 4. Identified 34 Staphylococcus sp. isolates were sensitive to Cefoxitin, Ciprofloxacin, Vancomycin, Tigecycline and Phosphomycin (100%) and Linezolide (85.2%); resistant to Clindamycin (100%) and Tetracycline (82.4%). S. aureus isolates were detected resistant to Cefoxitin, Vancomycin, Tigecycline, Phosphomycin and Clindamycin (100%) and Tetracycline (85.8%). It was determined that Coagulase Negative Staphylococci isolates were resistant to Cefoxitin, Vancomycin, Tigecycline and Fosfomycin (100%), Ciprofloxacin (92.5%), Linezolide (88.8%), Tetracycline and Trimetoprim/Sulfametoxazole (81.5%) and Clindamycin (100%). S. simulans isolates were resistant to clindamycin (100%), Teicoplanine (90%) and other 11 antibiotics (90%) used in this research. S. arlettae isolate was resistant to Clindamycin and Teicoplanin (100%), susceptible to other antibiotics. Clindamycin was found to be 100% resistant to S. epidermidis isolates were found resistant to Clindamycin (100%) and susceptible to the other 12 antibiotics (75-100%) used in this study. S. saprophyticus isolates were resistant to Clindamycin (100%) and susceptible to other 8 antibiotics (100%). S. chromogenes and >S. hyicus isolates (n = 2) were found to be resistant to Clindamycin and Teicoplanin (100%).
Table 3: CLSI standard minimal inhibitory concentration (MIC) values (CLSI, 2017)
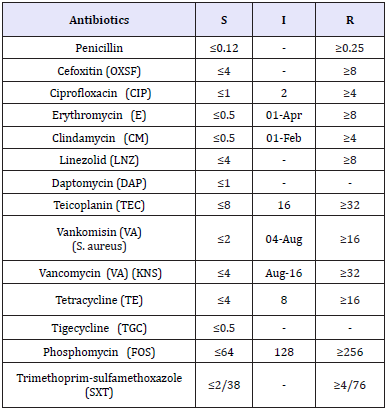
Table 4: The result of antibiogram in this study
Discussion
S. aureus is the most common bacterial cause of chronic mastitis. Although some cattle catch clinical mastitis after calving, infection is usually subclinical and causes an increase in somatic cell counts without noticeable changes in the milk or mammary lobes. Once established, S. aureus infection does not respond to antibiotic therapy and infected cattle must be removed from the herd [13,14]. Vaious isolation rates in different studies for the identification of microorganisms that cause mastitis has been found in Turkey. Türütoğlu et al. [15] reported 28.1% S. aureus and 23.1% S. epidermidis in the Marmara region, Kuyucuoğlu and Uçar [5] detected 40.1% S. aureus in the Afyon region, Ergün et al. [16] found 42.4% CoNS and 25.1% S. aureus, Gürtürk et al. [17] have isolated 41% of Staphylococcus species in Van and its region. Yıldız [18] identified 6 (31.58%) S. aureus, 5 (26.32%) S. epidermidis in clinical mastitis milk samples.
In studies conducted abroad, Tenhagen et al. [19] identified 9.1% CoNS and 5.7% S. aureus in Germany, Gianneechini et al. [20] reported 62.8% S. aureus and 7.4% CoNS in Uruguay, Pitkälä et al. [21] identified 49.6% CoNS and 10.2% S. aureus in Finland, Workineh et al. [22] identified 40.5% S. aureus and 16.5% CoNS in Ethiopia. Lüthje and Schwarz (2006) analysed antimicrobial agents of Coagulase Negative Staphylococci and Macrolide and Linkosamide (ML) resistance for the control of pathogens associated with bovine mastitis. In total, 298 CNS isolates were collected from subclinical mastitis cattle in Germany between 2003 and 2005. S. chromogenes (99 isolates, 32.2%), S. simulans (69 isolates, 23.2%), S. epidermidis (35 isolates, 11.7%), S. xylosus and S. haemolyticus, 9.4%). In addition, S. capricius, S. cohnii, S. hominis (2 isolates from each) and single isolate Staphylococcus caprae (1 isolate), S. sciuri (8 isolates), Staphylococcus equorum (6 isolates), S. saprophyticus, S. arlettae and Staphylococcus gallinarum species were identified.
Pankaj et al. [23] investigated the etiologic factors of Murrah cows with mastitis. A total of 326 milk samples were taken from 82 healthy cattle. Analyses have revealed 44 organisms as a result. 15.90% of them were Coagulase Positive Staphylococci, 47.72% were Coagulase Negative Staphylococci, 25% were Streptococcus dysgalactiae, 9.09% were Streptococcus agalactiae, 2.27% were Streptococcus uberis and 13.63% Staphylococcus spp. and Streptococcus spp. S. aureus and Staphylococcus haemolyticus was detected as major isolates, followed by S. epidermidis, S. simulans, S. hyicus, Staphylococcus pasteuria, Staphylococcus saprophyticus subsp. saprophyticus, S. arlettae and S. gallinarum isolates.
The phenotypic differentiation of Coagulase Positive Staphylococci species presents a difficult situation due to the lack of specific biochemical markers. To overcome this problem, the use of automatized tools has become routine in human and veterinary microbiology. Nevertheless, phenotypic analyses are relatively time consuming and most importantly, difficult to analyse results [24]. In this study, identification of S. aureus and other coagulase negative species isolated by conventional and commercial biochemical identification methods was verified by VITEK 2, VITEK (MALDI TOF) MS analyses. Automated systems have proven to be an effective tool to distinguish Staphylococcal species.
β-lactam antibiotics have been used for many years to treat staphylococcal infections. The unconscious use of the β-lactam group of antibiotics has also led to the development of resistance to these antibiotics. When the literature data are examined; with regard to Staphylococci isolates, Penicillin resistance is in the ratio of 59-88% and Amoxicillin resistance is 29-78% [5,25,26] for Turkey’s survey conducted in different regions. In this study, β-lactam group for Staphylococci isolated from mastitis milk were tested in antibiotic susceptibility tests; Penicillin is 35.3% resistant and Cefoxitin is 100% sensitive. In experiments with other antibiotic groups, strains were 100% sensitive Vancomycin, Tigecycline and Fosfomycin. The Staphylococcus strains isolated in our study showed 100% resistance to clindamycin. The high rate of Vancomycin susceptibility is considered due to the very limited use of vancomycin-containing preparations [27,28].
Conclusion
In addition to the standard biochemical procedures we use for identification in our study, the use of other automated systems has been shown to produce similar results compared to the factors isolated in mastitis studies in our country. Therefore, it has been demonstrated that the reliability of proteomic methods such as MALDI-TOF MS can be verified and applied. Thanks to our research, the isolation and identification of the pathogens in the clinic type mastitis cases, which are very important for milk production, with fast and reliable methods, will be shed light on future studies for the genetic and the peripheral of the disease. Future work may be effective in determining the diagnosis and treatment policies of the disease for our country. It should be taken into consideration that different antibiotic susceptibilities may also be detected in our studies when both antibiotic treatment and S. aureus and other Coagulase Negative Staphylococci species are present.
Acknowledgement
This research was supported by the Adnan Menderes University Scientific Research Projects Committee (code No. VTF-17010).
References
- Waldvogel FA (2000) Staphylococcus aureus (Including Staphylococcal Toxic Shock). Mandell, Douglas and Bennet’s Principles and Practice of Infectious Diseases. Churchill Livingstone, New York, USA pp. 2069- 2092.
- Tünger A (2004) Staphylococcus aureus: Mikrobiyoloji, Patogenez ve Epidemiyoloji. Gram-Pozitif Bakteri İnfeksiyonları. Bilimsel Tıp Yayınevi, Ankara p. 9-68.
- Tünger A, Çavuşoğlu C, Korkmaz M (2005) Mikrobiyoloji. Asya Tıp Kitapevi, İzmir, p. 6.
- Kırkan Ş, Göksoy EÖ, Kaya O (2005) Identification and Antimicrobial Susceptibility of Staphylococcus aureus and Coagulase Negative Staphylococci From Bovine Mastitis in the Aydın Region of Turkey. Turk J Vet Anim Sci 29: 791-796.
- Kuyucuoğlu Y, Uçar M (2001) Afyon Bölgesi Süt İneklerinde Subklinik ve Klinik Mastitislerin Görülme Oranları ve Etkili Antibiyotiklerin Tespiti. Veteriner Hekimleri Mikrobioloji Dergisi 1: 19-24.
- Türütoğlu H, Erçelik S, Öztürk D (2006) Antibiotic Resistance of Staphylococcus aureus and Coagulase-Negative Staphylococci isolated From Bovine Mastitis. Bull Vet Inst Pulawy 50: 41-45.
- Holland RD, Wilkes JG, Rafii F, Sutherland JB, Persons CC, et al. (1996) Rapid Identification of Intact Whole Bacteria Based on Spectral Patterns Using Matrix Assisted Laser Desorption/Ionization with Time-Of-Flight Mass Spectrometry. Rapid Commun Mass Spectrom10(10): 1227-1232.
- Wieser A, Schneider L, Jung J, Schubert S (2012) MALDI-TOF MS in Microbiological Diagnostics of Microorganisms and Beyond. Appl Microbiol Biotechnol 93(3): 965-974.
- Van Veen SQ, Claas EC, Kuijper EJ (2010) High-throughput Identification of Bacteria and Yeast by Matrix-Assisted Laser Desorption İonization- Time Of Flight Mass Spectrometry in Conventional Medical Microbiology Laboratories. J Clin Microbiol 48(3), 900-907.
- Eigner U, Holfelder M, Oberdorfer K, Betz-Wild U, Bertsch D, et al. (2009) Performance of a Matrix- Assisted Laser Desorption Ionizationtimeof- Flight Mass Spectrometry System for the Identification of Bacterial Isolates in the Clinical Routine Laboratory. Clin Lab 55(7-8): 289-296.
- Seng P, Drancourt M, Gouriet F, La Scola B, Fournier PE, et al. (2009) Ongoing Revolution in Bacteriology: Routine Identification of Bacteria by Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry. Clin Infect Dis 49(4): 543–551.
- Winn W, Allen S, Janda W, Koneman E, Procop GW,et al. (2006) Gram- Positive Cocci. Koneman’s Color Atlas and Textbook of Diagnostic Microbiology. Lippincott Williams Wilkins, Baltimore, pp. 623-671.
- Petersson-Wolfe CS, Mullarky IK, Jones GM (2010) Staphylococcus aureus Mastitis: Cause, Detection, and Control. Virginia Cooperative Extension pp. 404-229.
- Paterson GK, Morgan FJE, Harrison EM, Peacock SJ, Parkhill J, et al. (2014) Prevalence and Properties of mecC Methicillin-Resistant Staphylococcus aureus (MRSA) in Bovine Bulk Tank Milk in Great Britain. J Antimicrob Chemother 69(3): 598-602.
- Türütoğlu H, Ateşoğlu A, Salihlioğlu H, Öztürk M (1995) Marmara Bölgesi Süt İneklerinde Mastitise Neden Olan Etkenler. Pendik Veteriner Mikrobiyoloji Dergisi 26: 125-137.
- Ergün Y, Aslantaş Ö, Cantekin Z, Doğruer G (2004) Hatay İlindeki Aile Tipi Süt Sığırcılığı İşletmelerinde Subklinik Mastitislerin Epidemiyolojisi. Vet Bil Derg 20(4): 25-28.
- Gürtürk K, Boynukara B, Ekin İE, Gülhan T (1998) Van ve Yöresindeki İneklerde Subklinik Mastitisin Etiyolojisi Üzerine Bir Çalışma. Y Y Ü Vet Fak Derg 9(1-2): 1-4.
- Yıldız A (2003) Laktasyondaki Subklinik ve Klinik Mastitisli Sütçü İneklerde Lincomycin- Neomycin Kombinasyonuyla Meme İçi Tedavinin Etkinliği. Fırat Üniversitesi Sağlık Bilimleri Dergisi 7(1): 65-69.
- Tenhagen BA, Köster G, Wallmann J, Heuwieser W (2006) Prevalence of Mastitis Pathogens and Their Resistance Against Antimicrobial Agents in Dairy Cows in Brandenburg, Germany. J Dairy Sci 89(7): 2542-2541.
- Gianneechini R, Concha C, Rivero R, Delucci I, Moreno López J (2002) Occurrence of Clinical and Subclinical Mastitis in Dairy Herds in The West Littoral Region in Uruguay. Acta Vet Scand 43(4): 221-230.
- Pitkälä A, Haveri M, Pyörälä S, Myllys V, Honkanen-Buzalski T (2004) Bovine Mastitis in Finland 2001 Prevalence, Distribution of Bacteria, and Antimicrobial Resistance. J Dairy Sci 87(8): 2433-2441.
- Workineh S, Bayleyegn M, Mekonnen H, Potgieter LN (2002) Prevalence and Aetiology of Mastitis in Cows From Two Major Ethiopian Dairies. Trop Anim Health Prod 34(1): 19-25.
- Pankaj SA, Chhabra R, Anshu Sharma, Sindhu N (2013) Sub-clinical Mastitis in Murrah Buffaloes with Special Reference to Prevalence, Etiology and Antibiogram. Buffalo Bulletin 32(2): 107-115.
- Sasaki T, Tsubakishita S, Tanaka Y, Sakusabe A, Ohtsuka M, et al. (2010) Multiplex-PCR Method for Species Identification of Coagulase-Positive Staphylococci. J Clin Microbiol 48(3): 765-769.
- Şahin M, Çolak A, Otlu S, Aydın F, Genç O, et al. (1997) Kars Yöresi İthal Simental İneklerinde Subklinik ve Klinik Mastitislerin Görülme Oranı ve Etkili Antibiyotiklerin Belirlenmesi. Kafkas Üniversitesi Veteriner Fakültesi Dergisi 3(1): 49-55.
- Akan M, Kökçü L, Öncel T, Ekem S (2001) Mastitislerden izole Edilen Stafilokok Suşlarının Beta Laktamaz Aktivitesi ve Bazı Antibiyotiklere Duyarlılıkları. Veteriner Hekimleri Mikrobiyoloji Dergisi 2: 31-34.
- CLSI National Committee for Clinical Laboratory Standarts (M11) (2017) Performance Standards for Antimicrobial Susceptibility Testing. Vol. 27th Informational Supplement, Wayne.
- Lüthje P, Schwarz S (2006) Antimicrobial Resistance of Coagulase Negative Staphylococci From Bovine Subclinical Mastitis with Particular Reference to Macrolide-Lincosamide Resistance Phenotypes and Genotypes. J Antimicrob Chemother 57(5): 966-969.
© 2018 Kırkan Sukru. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






