- Submissions

Full Text
Approaches in Poultry, Dairy & Veterinary Sciences
Isolation of Major Pathogens from Cattle with Subclinical Mastitis and Determination of Antibiotic Susceptibilities
Parin Ugur1*, Kirkan Sukru1, Ozdemir Fatma Özge2 and Yuksel Hafize Tugba1
1 Department of Microbiology, Adnan Menderes University, Turkey
2 Department of Microbiology, Institute of Health Sciences, Turkey
*Corresponding author: Parin Ugur, Department of Microbiology, Faculty of Veterinary Medicine, Adnan Menderes University, Aydin, Turkey, 09016
Submission: March 23, 2018; Published: April 19, 2018

ISSN: 2576-9162 Volume3 Issue3
Abstract
Bacterial factors of bovine mastitis have two groups according to origin and transmission routes. The primary contagious agents are S. aureus, Streptococcus sp. and E. coli. These factors are spread widely, especially with handling and milking units. The scope of this study was to determine the isolation and antibiotic susceptibility of S. aureus, Streptococcus sp. and E. coli, which are very important for milk production and frequent agents of subclinical mastitis. In our study, 138 samples of dairy cows were taken aseptically from Aydin province with subclinical mastitis case. In conclusion, 102 (18.5%) S. aureus, 21 (3.8%) Streptococcus sp., and 15 (2.7%) E. coli were isolated from the 552 milk samples examined. After PCR with specific primers in the genotypic identification, all isolates were confirmed about bacterial identification. S. aureus, Streptococcus sp., and E. coli isolates were susceptible to antibiotics of amoxicillin-clavulanic acid, oxytetracycline, cefoperazone and cephalexin-kanamycin.
keywordsSubclinical mastitis; S. aureus, Streptococcus sp; E. Coli; PCR; Antibiogram
Introduction
Mastitis, commonly known as inflammation of the mammary glands, is usually associated with microbial infections [1] and is defined as mammary gland inflammation, not as an Intra Mammary (IMI) Infection. Mastitis, an endemic disease in dairy cattle farms around the world, has influence on the loss of milk production. Moreover, mastitis has a direct impact on milk quality, both technically and hygienically. Mastitis treatment is therefore to improve milk quality and the productivity of milk production, there by making milk production more sustainable [2]. Mastitis continues to be the most expensive disease in the world, in clinical or sub clinical form, found in dairy farms [3].
In the spontaneous diseases, the mastitis agents reach the mammary tissue in many ways. These infections emerge by fragile milk ducts, nipple wounds, and infections in the upper respiratory organs, which come into contact with the mammary tissue via blood. There is a significant decrease in milk production both in clinical and sub clinical mastitis. Even after mastitis has been cured in loss of milk production, the yield is not as good as that of the animal; even if the animal recovers [2]. The most common bacterial agents of mastitis are Streptococci (5.43%-20.35%), Coagulase Negative Staphylococci (CNS) (2.86-58.15%) and members of Enterobacteriaceae family (8.47%). Staphylococcus aureus alone accounts for 19.97-65.0% of mastitis cases. Coagulase Negative Staphylococci, which are considered to be the causative pathogens in the development of mastitis now adays, are of great interest in veterinary medicine. CNS is not as pathogenic as other mastitis agents. However, it can cause permanent infections, although it often occurs sub clinically. The bacterial disease leads to an increase in SCC and a drop in milk quality [1].
In recent years, pathogens that produce antibiotic-resistant mastitis have become an increasing concern worldwide. To administer management practices to prevent and treat mastitis, it is necessary to know not only which pathogens cause mastitis, but also which antibiotics are susceptible to these pathogens. The presence of various bacteria in mastitis has been identified in studies conducted, and the distribution of specific infectious agents has been influenced by cattle breeding, lactation stage, milk yield and previous lactation count. Dairy cattle have been shown to be susceptible to mastitis pathogens more than beef cattle.
Today, information about mastitis and optimal technical management and treatment are available. The prevalence of mastitis is lowered with the development of protective measures at the farm level and the correct antibiotic treatment. Antibiotic use is also reduced using prudent antibiotic administration methods.
In appropriate practices can be defined as the reduction of misuse and, where appropriate, the increased use of additional treatments such as alternative medicines. However, producers are still applying inappropriate antibiotics [4].
In this study, isolation of S. aureus, Streptococcus sp. and Escherichia coli strains, determination of antibiotic susceptibility were aimed in milk samples taken from 138 animals showing sub clinical mastitis problem in Aydın province. It is predicted that the results of the study may be useful in the diagnosis and treatment of mastitis cases that may occur in our future. However, there is information in our study results regarding comparing the characteristics of mastitis-affecting organisms of different regions of Turkey and presenting regional data for future studies.
Materials and methods
Specimen collection
In this study, 552 milk samples were taken from 138 dairy cows with subclinical mastitis from a dairy cattle enterprise in the Incirliova district, Aydin province within three months period (2017 May-August). The California Mastitis Test (CMT) was used to detect subclinical mastitis cow milk. All of the Holstein breed cows in the study were in lactation but did not receive antibiotic therapy within the last month. The age scale of the cows was between 3 and 11.
Primer pairs
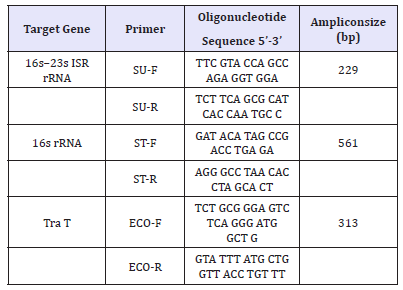
The primer pairs for identification of E. coli, S. aureus, and Streptococcus sp. were shown in (Table 1).
Table 1:Target gene, Oligonucleotide Primer Sequences and Amplicon Sizes (Pradhan et al., 2011)
Collection of specimen
Before collecting samples from animals, subclinical mastitis was detected by CMT. All mammary nipples were cleansed with 70% alcohol. After a few milliliters of milk were poured off, 10 ml of milk sample were collected from 4 mammary lobes. The samples were sent to the Routine Diagnostic Laboratory of Department of Microbiology, Department of Veterinary Medicine of Adnan Menderes University under cold chain for isolation.
S. aureus and Streptococcus sp. isolation : Milk samples were inoculated onto blood agar containing 5% sheep blood and allowed to incubate for 48 hours at 37 °C. Hemolytic colonies on blood agar were then inoculated to Tryptic Soy Agar (TSA) to recover the pure culture, and allowed to incubate at 37°C for 24 hours. Gram staining was applied to determine the Gram(+), and Gram(-) isolates in order to identify the bacterial strains S. aureus, Streptococcus sp. and Escherichia species. Catalase test was performed for differentiation of Staphylococcus and Streptococcus sp. Catalase negative colonies were preliminarily identified as Streptococcus sp. Coagulase positive colonies were regarded as S. aureus. Gram(-) isolates were inoculated to Eosin Methylene Blue (EMB) agar and Mac Conkey agar, then incubated at 37°C for 24 h for identification of E. coli). The isolates with a greenish metallic sheen on EMB agar and lactose positive pink colored isolates were preliminarily identified as E. coli after IMVIC (Indole, Methyl red, Voges-Proskauer, Citrate) reactions.
Bacterial strains
Staphylococcus aureus ATCC 25923 (16s-23s ISR positive), E. coli ATCC® 25922 (tra T positive) and Streptococcus sp. ATCC ® 12392 (16s rRNA positive) were obtained from the manufacturer
Genotypic identification
DNA extraction from isolates was performed via DNA Extraction Kit (Fermentas®, Lithuania) according to manufacturer’s specifications. The concentrations of DNA obtained by extraction kit were determined with a micro-volume spectrophotometer (ProNano PN-913, Maestrogen®, Taiwan). PCR primers were synthesized by manufacturer highly divergent and species-specific regions of the DNA coding for 16S-23S ISR rRNA, 16S r RNA and Trat gene for the detection of Staphylococcus, Streptococcus sp. and E. coli respectively.
The primers were diluted in TE buffer to a final concentration of 1000 pmol/μl and stored at-80 ˚C as stock. The working concentration of 20pmol/μl was prepared with sterile filtered deionized water, aliquoted and stored at-20 ºC. PCR was performed in a Gene Amp PCR Systems 9700 (Applied Biosystems®) in a final volume of 25μl consisting of 5pmol each of the primer pair, 1 U of Taq DNA polymerase (Fermentas®), 25mM MgCl2, and 100μM each of the four deoxyribonucleotide triphosphates (Fermentas®) in 25mM Tris-HCl, pH 8.3. Initial denaturation was performed at 95 ˚C for 2 min before applying 35 cycles, each cycle with denaturation at 95 ˚C for 45 s, annealing 50 ˚C for 1min, and extension at 72 ˚C for 30sec. After the final cycle, one cycle of extension at 72 ˚C for 5min was carried out to complete the reaction. PCR assay was performed for each pathogen using known standard DNA and with 5pmol each of the specific primer pair described before [5]. Subsequently, PCR was performed with a mixture of standard DNAs with ten-fold serial dilutions of templates to rule out non specific amplification. The PCR products were loaded on agarose electrophoresis gel, and stained with three μl ethidium bromide. The bands were referenced to a Gene Ruler 100-bp DNA ladder (Fermentas®, Canada) and a 1-kb DNA ladder (Fermentas®, Canada) to size the amplicons.
Antibiotic susceptibility Test
Disc diffusion method was applied using Mueller-Hinton Agar (Merck Millipore®, Germany) to determine the antibiotic susceptibilities of isolates (CLSI, 2017). Selected antimicrobial agents used in the antibiotic susceptibility test were Penicillin G, Oxytetracycline, Cefoperazone, Neomycin, Neomycin-Bacitracin- Tetracyclin, Cephalexin-Kanamycin, and Amoxicillin-Clavulanic acid.
Results
Of the 552 milk samples, 102(18.5%) S. aureus, 21(3.8%) Streptococcus sp, and 15(2.7%) E. coli isolates were obtained. Also, 23(4.1%) Corynebacterium sp. and 77(13.9%) Lactobacillus sp. were the other bacterial species identified from samples. No bacterial growth was detected in 314(56.8%) specimens.
Genotypic identification results
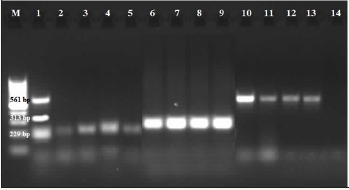
For genotypic identification, 16S-23S ISR(Intergenic Spacer Region) rRNA in S. aureus isolates, Streptococcus sp. 16S rRNA in E. coli isolates and Trat (complement resistance protein) genes were genotypically examined. After PCR with specific primers, S. aureus, Streptococcus sp. and E. coli positive PCR product bands were screened in lengths of 229, 561 and 313bp, respectively (Figure 1).
Figure 1:Gel electrophoresis image M: 100 bp Marker, 1: S. aureus, Streptococcus sp. and E. coli positive controls, 2-5: S. aureus positive samples, 6-9: E. coli positive samples, 10- 13: Streptococcus sp. positive samples, 14: Negative controls
Antibiotic susceptibility results
Table 2:The susceptibility rates of the relevant antibiotics NBT: Neomycin-Bacitracin-Tetracycline CFX/K: Cephalexin-Kanamycin N: Neomycin CFP: Cefoperazone P: Penicillin G AMC: Amoxycilline-Clavulanic Acid OT: Oxytetracycline

Antibiotic susceptibility rates of bacterial isolates as a test result are presented in Table 2.
Discussion
In this study, PCR method has overcome the restrictions for detection of multiple-bacterial subclinical mastitis. The test depends on multiplex PCR that can distinguish pathogens. PCR techniques are proficient for the location of pathogens at low fixation. Techniques including ribotyping are valuable for pathogen location. Along these lines, PCR based location of 16S rRNA or 23S rRNA primers have been effectively connected to the distinguishing of numerous microorganisms. The favourable position of PCR is its affectability as a couple of picogram amounts of nucleic acid is sufficient to distinguish the detection of various pathogens, permitting the confirmation of culture. Also, rRNA is available in numerous duplicates, which increases the specifity. Primers depicted here were turned out to be particular since on agarose gel just a single band was screened for each fragment apart from other ones, and no amplification was identified with negative controls. The procedure was applicable for multiplex PCR and regarded as specific for S. aureus, Streptococcus sp. and E. coli. Subclinical bovine mastitis is a costly and multi factorial disease for breeders due to reduced milk production, economic losses and untreated chronic mortality rates in the dairy industry. Mastitis is often caused by a variety of infectious agents (usually bacteria), along with physical and chemical agents. More than 120 different microorganism species, including bacterial, viral and fungal, have been isolated from mastitis cows’ milk. The most common causes of bacterial mastitis are E. coli, S. aureus, Streptococcus sp. and Enterococcus faecalis [6, 7].
The scope of this study is to identify isolated strains by phenotypic and molecular methods. Obaidat et al. [8] conducted a research to identify significant anti microbial agents and resistance against E. coli from cattle in Jordan. A total of 520 E. coli isolates from 43 farms were investigated. 12 antimicrobials were tested for resistance to mastitis in 51.2% of the samples and the most commonly used antimicrobials were oxy tetracycline and streptomycin. E. coli isolates showed high resistance to streptomycin (47.5%), tetracycline (45.4%) and ampicillin (34.2%). Less than 10% of the isolates were resistant to kanamycin, gentamicin, ciprofloxacin and ceftriaxone. In general, 64.6% and 37.1% of E. coli isolates were resistant to ≥1 antimicrobial and multidrug resistance (resistance to ≥3 antimicrobial classes), respectively. The isolates exhibited an antimicrobial resistance profile of 107, and E. coli antibiotic resistance was found to be high in cows in Jordan [8, 9] collected 489 milk samples from the breast lobes of 273 dairy cows and 393 (80.37%) samples showed bacterial isolation while 55 (11.25%) samples showed no breeding. Also, 8.38% of the specimens were found to have 3 or more bacterial species in the medium, suggesting that the samples were contaminating. Eightynine (20.10%) bacterial culture positive strains were found to be E. coli strains, and the incidence of E. coli from different farms was between 15.7% and 25%.
In the antimicrobial susceptibility tests of E. coli isolates, they were found to have resistance to sulfamethoxazole 100%, sulfamerazine 98.73%, tiamfenicole 93.67% and oxytetracycline 79.75%. Also, they were found to have high resistance to colistin 49.37%, ceftiofur 22.91%, cefazolin 31.65%, azithromycin 31.65%, ampicillin 25.32% and tetracycline 25.32%, and low resistance to doxycycline, chloromycetin, florfenicol, amoxicillin, ciprofloxacin, streptomycin, and amikacyn. Although there are antimicrobial agents that are effective against the isolates ofloxacin, gentamicin, and enrofloxacin, they have been reported to be resistant to these antimicrobial agents.
Milk samples from cows with acute clinical mastitis were collected using the Vet Path program in nine countries, and 934 isolates were examined for subsequent antimicrobial resistance tests between 2009 and 2012. Milk samples were taken from cows with acute clinical mastitis from Belgium, the Czech Republic, Denmark, France, Germany, Italy, the Netherlands, Spain and the United Kingdom. When the samples were taken, 79.1% of the animals were found not to use antibiotics for at least three weeks.
The remaining 20.9% was treated as unknown. Only one sample per year was taken from each run. The samples were sent to the central laboratories for identification by microbiological and biochemical tests according to gender and species level. Here, the MIC values of all samples were determined as twofold dilutions, by CLSI standards, amoxicillin-clavulanic acid, cefquinome, ceftiofur, cephalin, cephapirin, cefalonium, cloxacillin, penicillin G, enrofloxacin, marbofloxacin, erythromycin, tylosin, kanamycin, cephalexin / kanamycin, neomycin, and tetracycline for 16 combinations of antibiotics. MIC values against penicillin G, cloxacillin, erythromycin, and tylosin against Gram (-) bacteria were not reported due to intrinsic resistance. In total 934 cases of acute mastitis, 207 E. coli, 87 Klebsiella sp., 192 S. aureus, 165 CNS, 188 S. uberis and 95 S. dysgalactiae were isolated. Resistance to five antibiotics in studies between VetPath 2002-2006 and VetPath 2009-2012 was compared regarding the three pathogens seen in both studies. The resistance of E. coli, S. aureus and S. uberis compared to 2009-2012 for 2002-2006, the penicillin resistance of S. aureus was significantly lower for 2009-2012. There was no statistically significant difference between the two periods. In contrast, the resistance of S. uberis to tetracycline has increased significantly between 2009 and 2012 [10].
Suleiman et al. [11] surveyed to determine the prevalence of sub clinical mastitis and the antibacterial susceptibility patterns of isolated mastitis pathogens in cows in different farms in Zanzibar, between January and July 2014. To test the susceptibility ratio of common isolated bacteria, disc diffusion technique was used, 1664 mammary lobes were scanned, and 1648 samples were sent to the laboratory for bacterial culture after CMT. According to the results of CMT, the general subclinical mastitis prevalence was determined as 28.6% in mammary lobes, 48.8% in individual level and 64.7% in farm level respectively.
The isolates were S. aureus (36.8%), P. aeruginosa (17.8%), S. epidermidis (16.1%), Klebsiella sp. (9.5%), Micrococcus sp, (6.3%) and E. coli (4.9%). As a result, subclinical mastitis was observed in cows and high level in farms with both infectious and environmental bacterial pathogens [11, 12] reported that in the study of 260 sick cows which were clinically treated with oxytetracycline, enrofloxacin, gentamycin, penicillin, ceftriaxone, cefotaxime, ceftriaxone/tazobactam, amoxicilline-clavulanic acid, cefoperazone, ampicilline sulbactam, neomycin, ciprofloxacin, ceftizoxime and tylosin antibiotics were used. According to the results of the research, the incidence of clinical mastitis was found to be 11.5%. In the bacteriological examination of the milk of the animals that had previously had mastitis, 14(60.87%) S. aureus, 2(8.69%) S. dysgalactiae, 3(13.04%) Coagulase Negative Staphylococci, 1(4.35%) S. uberis and 3(9.09%) were identified as other coliform bacteria. Staphylococcus sp., Streptococcus sp. and E. coli isolates were highly susceptible to enrofloxacin and gentamicin followed by amoxicillin-clavulanic acid, ampicilline sulbactam, ceftriaxone-tazobactam. Koçyiğit et al. [13] collected samples from 774 mammary lobes of 195 Holstein, Swiss Brown and Simmental dairy cows tested by CMT in their research and microbiologically assessed milk samples taken from 125 mammary lobes of 100 cows that had a CMT positive reaction in at least one lobe.
The CMT positive cow rate was found to be 51.28%, while 63% of their milk samples had bacteriological growth. E. coli (28.9%), Candida sp. (24.21%), S. uberis (19.53%), Coagulase Negative Staphylococci (19.53%), S. aureus (3.9%) and Proteus sp. (2.34%) were identified from samples. Also, no statistically significant difference was found between the number of lactations of CMT positive cows on farms. In other words, farm location, age, lactation number/period, localization of mammary lobes did not affect microbiological results. In microbiologically positive samples, microorganisms were found to be sensitive to amoxicilline clavulanic acid and oxytetracycline, while resistant to gentamicin, ceftiofur, enrofloxacin and cefquinom [13].
Rüegsegger et al. [14] evaluated the antimicrobial resistance of 3954 milk samples isolated from samples collected from animals with subclinical and clinical mastitis diagnoses from various regions of Switzerland between 2011 and 2013. Staphylococcus spp, 598
(15%) coliform, 490(12%) S. aureus and 270(7%) Enterococcus sp. were detected in the obtained samples, and 213(5%) S. dysgalactiae were isolated and the prevalences of Mannheimia haemolytica, Pasteurella sp., Proteus sp., P. aeruginosa, Serratia sp., S. agalactiae and other Streptococci were determined to be less than 1%. Nine antimicrobial drugs (amoxicilline-clavulanic acid, ampicilline, cefoperazone, gentamycin, lincomycin, oxacillin, penicillin, polymyxine, spiramycin) were tested for sensitivity. Test results showed that S. aureus isolates were resistant to penicillin, while 1.2% isolate showed the highest resistance against amoxicilline clavulanic acid. Of 598 coliform isolates, 29(4.9%) were resistant to gentamicin, 270(45.1%) were resistant to ampicilline, and 66(11%) were resistant to amoxicilline clavulanic acid. In 3954 isolates of mastitis, resistance was found to be 86% for polymyxine, 64.7% for oxacillin, 53.7% for lincomycin, 45.5% for gentamicin, 39.2% for penicillin, 27.0% for spiramycin and 26.7% for ampicilline. Cefoperazone (92%) and amoxycilline-clavulanic acid (97.4%) were the most sensitive. S. uberis, S. dysgalactiae and S. aureus had the highest antimicrobial susceptibility values of 99.6%, 100% and 98.8% for amoxicilline-clavulanic acid, respectively. Total resistance level against gentamicin was 45.5%, penicillin 39.2% and ampicilline 26.7%.
The highest resistance levels were polymyxine (86.0%), oxacillin (64.7%), and lincomycin (53.7%). Thus, resistance to at least one of the antimicrobial agents has been demonstrated. A total of 256 milk samples from 61 animals were collected from 86 straws during the lactation period in the Sarikamis district of Kars. Of these samples, 72(91.1%) were subclinical, and 7(8.9) samples were related to clinical mastitis mammary lobes. When the microbiological examination of the milk was carried out, 67 isolates were detected from samples. As a result of the CMT test, the subclinical mastitis ratio in cows was found to be 22.3%, and the clinical mastitis rate was found to be 1.5%. It has been determined in a study that a significant portion of mastitis cases was subclinical. Of the 47 mastitic milk, Staphylococcus sp. and Micrococcus sp. Streptococcus sp. and 4 were lactose positive and metallic reflections colonies on EMB agar. Staphylococcus sp., isolated and identified from mastitis milk [15], Antibiotics against staphylococci were determined to be amoxicilline-clavulanic acid, cloxacillin, enrofloxacin, and vancomycin, which are the most effective antibiotics against staphylococci.
In our study, 102 (18.5%) S. aureus, 21 (3.8%) Streptococcus sp., and 15 (2.7%) E. coli were isolated from the 552 milk samples examined. After PCR with specific primers in the genotypic identification, all isolates were identified as S. aureus, Streptococcus sp. and E. coli. The obtained S. aureus, Streptococcus sp. and E. coli isolates were susceptible to antibiotics of amoxicillin-clavulanic acid, oxytetracycline, cefoperazone and cephalexin-kanamycin. The resulting data were determined by the studies carried out.
Conclusion
Cattle mastitis is a standout amongst the most monetarily critical ailments that influence the dairy industry. Billions of dollars are lost all over the world due to decreased quality and amount of drain delivered. In spite of the fact that the mastitis is of multietiological, the high level of misfortune is because of the three noteworthy bacterial pathogens, Staphylococcus sp., Streptococcus sp., and E. coli. Mastitis, if distinguished at subclinical level can be controlled by antimicrobial treatment. Diagnosis of subclinical mastitis and the causative agent is essential for under taking preventive measures. Customary methodology for identified proof of pathogens of mastitis is worked serious, and the greater part of the conventional tests is not intended to distinguish dominant veterinary pathogens. Location of mastitis at sub-clinical form needs tests that are exceedingly sensitive and PCR based tests can meet this necessity [16].
However, a few of these accessible techniques still need enhancement of the way of culture, which is tedious. Additionally to recognize the pathogen the PCR responses are to be conveyed independently for various pathogens. Molecular identification is an elective strategy, be that as it may, the similarity of the primer sets is essential for the response to work productively with high affectability of identification. Mastitis is an important mammary gland disease caused by microorganisms. Due to mastitis, big economic losses occur every year. The economic losses caused by the disease are not only limited to a reduction in milk yield but also include expenditures such as the treatment of the disease and the removal of diseased animals. Deterioration in the quality of the milk, which is an important food source, also has negative consequences for public health. It is expected that the results obtained in this study past will be beneficial for future studies.
Acknowledgement
This research was granted by Adnan Menderes University Scientific Research Projects Unit Aydın, Turkey (Project Grant Code: VTF-17029).
References
- Klimiene I, Virgailis M, Pavilonis A, Siugzdiniene R, Mockeliunas R, et al. (2016) Phenotypical and Genotypical Antimicrobial Resistance of Coagulase-Negative Staphylococci Isolated From Cow Mastitis. Pol J Vet Sci 19(3): 639-646.
- Hogeveen H, Osteras O. Mastitis (2005) Managemenet in an Economic Framework. In: Hogeveen (Ed) Mastitis in Dairy Production H. Wageningen Academic Publishers Wageningen, Netherlands, pp. 41-52.
- Supre´ K, Lommelen K, Meulemeester LD (2014) Antimicrobial Susceptibility and Distribution of Inhibition Zone Diameters of Bovine Mastitis Pathogens in Flanders, Belgium. Veterinary Microbiology 171(3-4): 374-381.
- Poizat A, Bonnet-Beaugrand F, Rault A, Fourichon C, Bareille N (2017) Antibiotic Use by Farmers to Control Mastitis as İnfluenced by Health Advice and Dairy Farming Systems. Prev Vet Med 146: 61-72.
- Pradhan P, Gopinath SM, Reddy GR, Dechamma HJ, Suryanarayana VVS (2011) Detection of Major Pathogens in Bovine Sub-Clinical Mastitis by Multiplex PCR Directly From Milk Samples in Presence of an Internal Control. Indian Journal of Fundamental and Applied Life Sciences 1(4): 209-218.
- Bes M, Guerin-Faublee V, Meugnier H, Etienne J, Freney J (2000) Improvement of the Identification of Staphylococci isolated from Bovine Mammary Infections Using Molecular Methods. Vet Microbiol 71(3-4): 287-294.
- Whitehead TR, Cotta MA (2000) Development of Molecular Methods for Identification of Streptococcus bovis From Human and ruminal Origins. FEMS Microbiol Lett 182(2): 2370-240.
- Obaidat MM, Salman AB, Davis MA, Roess AA (2018) Major Diseases, Extensive Misuse, and High Antimicrobial Resistance of Escherichia coli in Large- and Small-Scale Dairy Cattle Farms in Jordan. J Dairy Sci 101(3): 2324-2334.
- Zhang D, Zhang Z, Huang C, Gao X, Wang Z, et al. (2018) The Phylogenetic Group, Antimicrobial Susceptibility, and Virulence Genes of Escherichia coli From Clinical Bovine Mastitis. J Dairy Sci 101(1): 572-580.
- Jong A, Garch FE, Simjee S, Moyaert H, Rose M, et al. (2018) Monitoring of Antimicrobial Susceptibility of Udder Pathogens Recovered From Cases Of Clinical Mastitis in Dairy Cows Across Europe: VetPath results. Vet Microbiol 213: 73-81.
- Suleiman TS, Karimuribo ED, Mdegela RH (2018) Prevalence of Bovine Subclinical Mastitis and Antibiotic Susceptibility Patterns of Major Mastitis Pathogens Isolated in Unguja Island of Zanzibar, Tanzania. Trop Anim Health Prod 50(2): 259-266.
- Bhat AM, Soodan JS, Singh R, Dhobi IA, Hussain T,et al. (2017) Incidence of Bovine Clinical Mastitis in Jammu Region and Antibiogram of İsolated Pathogens . Vet World 10(8): 984-989.
- Koçyiğit R, Yılmaz O, Özenç E, Uçar M (2016) Effect of Some Risk Factors on Subclinical Mastitis in Dairy Cows. Kocatepe Vet J 9(3): 185-193.
- Rüegsegger F, Ruf J, Tschuor A, Sigrist Y, Rosskopf M, (2014) Antimicrobial Susceptibility of Mastitis Pathogens of Dairy Cows in Switzerland. Schweiz Arch für Tierheilkd 156(10): 483-488.
- Sevinti DA, Şahin M (2009) Sığır Mastitislerinden İzole Edilen Stafilokok Suşlarının Beta-Laktamaz Aktivitesi ve Bazı Antibiyotiklere Karşı Duyarlılıklarının Saptanması. Vet Bil Derg 1(2): 23-28.
- Clinical and Laboratory Standards Institute (CLSI) Performans Standards for Antimicrobial Ausceptability Testing; 27th Informational Supplement 2017, M100-S26
© 2018 Parin Ugur. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






