- Submissions

Full Text
Associative Journal of Health Sciences
Comparison of Different Sampling Methods for Diagnosis of COVID-19
Eman Arpida1,2, Marwa Karghol1,2, Mawada Alroog2, Omayma Suweelim1,2, Sarah Alazoumi1,2, Ali Elarabi1,2 and Abdulwahab Kammon1*
1National Research Center for Tropical and Transboundary Diseases (NRCTTD), Alzintan, Libya
2Department of Microbiology, Faculty of Science, Alzintan University, Alzintan, Libya
*Corresponding author: Abdulwahab Kammon, National Research Center for Tropical and Transboundary Diseases (NRCTTD), Alzintan, Libya
Submission: November 16, 2022;Published: December 02, 2022

ISSN:2690-9707 Volume2 Issue2
Abstract
A total of 49 COVID-19 positive cases were included and 17 out of them were selected randomly and monitored after 5 days to compare the availability of the virus based on Ct values of the genes (N and ORF1ab) in nasopharyngeal swabs, saliva and throat swabs. This study states that the best place to collect samples for COVID-19 diagnosis is the nasopharyngeal but saliva can be used in the diagnosis within the first 5 days in people who suffer from health problems that prevent taking a nasopharyngeal swab.
Keywords:COVID-19; Libya; Zintan; Nasopharyngeal; Saliva; Throat; N gene; ORF1ab gene
Introduction
SARS-COV-2 belongs to the genus Betacoronavirus, was discovered in Wuhan, China in December 2019 causing severe acute pneumonia. The World Health Organization (WHO) latterly gave the name Covid-19 [1].
Several studies have been conducted to diagnose the emerging SARS-COV-2 using real time RT-PCR (rRT-PCR). In the literature, there were differences of results based on the sampling types all over the world [2-6]. In Libya, scanty of information were published on COVID-19. The prevalence of COVID-19 in the same area of our study was reported by Kammon et al. [7] as 25.12% in which nasopharyngeal swab technique is adopted for the diagnosis of COVID-19.
rRT-PCR assay which firstly used for diagnosis of SARS-CoV-2 was developed at the Charité Institute of Virology in Germany and approved by the WHO on January 13, 2020 [8]. This assay targeted the E gene of the COVID-19. Later on, many assays were developed and validated targeting different genes of COVID-19 such as nucleoprotein (N) gene, open reading frame (ORF) gene, spike (S) gene, etc. Although there were no significant differences in the results across gene targets in many studies, significant differences were found in Ct values in commercial kits targeting all genes except S gene [9]. Pojul et al. [10] found that the N gene requires a longer period of days to become negative compared to the ORF gene, the percentage of the N gene was (12.68%), while the percentage of the ORF gene was (12.09%). The objective of the current study was to compare between different sampling methods for the diagnosis of COVID-19 based on the Ct values of both N and ORF1ab genes.
Discussion
In the current study, 49 COVID-19 positive cases were included for comparison. Three different samples (nasopharyngeal swab, throat swab and saliva) were collected from each case. Moreover, 17 out of the 49 positive cases were selected randomly and monitored after 5 days. The monitoring aimed to compare the availability of the virus based on Ct values and the gene amplification in each sample. The study was conducted during the time period from December, 2020 to April, 2021. This study was approved by Libyan National Committee for Biosafety and Biotechnology.
RNA was extracted from all different samples using automated system (NuActor, Bioditech, Korea). As per the manufacturer’s instructions, the samples are placed in the cartridge before it is placed in the NuActor device. The system then takes care of the rest to extract highly purified viral RNA automatically in about 12 minutes.
rRT-PCR commercial kit was used for the detection of SARSCoV- 2 nucleoprotein gene (N), open reading frame gene (ORF1ab) and human housekeeping (RNAse P) gene as an Internal Control (IC) (DaAn Gene, China). The reaction and program were conducted as per the manufacturer’s instructions using Azure Cielo 6 Real time PCR System (Azure Biosystems, USA). Samples with quantitation cycle values (Ct) ≤ 40 were considered positive for COVID-19.
Univariate statistical analyses using SPSS 26 computer program (SPSS Inc. Chicago, Illion, USA) was performed to determine the differences of Ct values between different sampling methods, using two different genes and time at first diagnosis and after 5 days.
The Ct values of N, ORF1ab and IC genes amplified from nasopharyngeal swabs, saliva and throat swabs collected from the 49 positive cases are shown in Table 1. All tested samples were positive for IC gene. This indicates the successful sampling for all methods used. The IC gene used in the current study is the housekeeping RNAse P which is a ribozyme expressed in many human tissues. In contrast to Skolimowska et al. [11] who found higher Ct values for housekeeping gene in saliva, the result of our study showed higher Ct values of IC gene in throat swabs, although it was not significantly different. A larger Ct value means more PCR cycles are needed for the amount of DNA or cDNA to reach the threshold allocated either manually by researcher or automatically by the machine, which indicates that there is a smaller amount of nucleic acid. However, it is previously reported that the buccal cavity had significantly more epithelial cells in buccal swabs than in saliva samples in both children and adults [12]. This could be attributed to the fact that saliva contains only sloughed-off epithelial cells. The most likely reason is that the direct scraping of the cheek enriches for such cells, whereas saliva will only contain sloughed-off cells. However, it would be interesting to study the relationship between cell types of the buccal cavity and expression of housekeeping genes. There are three buccal epithelial cell types of intermediate squamous cells, non-keratinous and keratinous superficial squamous cells [12].
Table 1:The Ct values of N, ORF1ab and IC genes amplified from different samples of 49 positive cases.
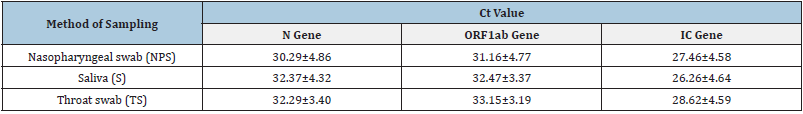
Values indicate means ± S.D.
The monitoring of the 17 positive cases revealed that all the samples were still positive after 5 days for both N and ORF1ab genes since the Ct values ≤40 is considered positive (Table 2). Statistically significant differences between sampling methods were observed. Nasopharyngeal swabs had significantly less Ct values in all tested genes as compared with other sampling methods at first diagnosis and even after 5 days (p˂0.01)
Table 2:Monitoring of 17 positive cases after 5 days of the first diagnosis of COVID-19.
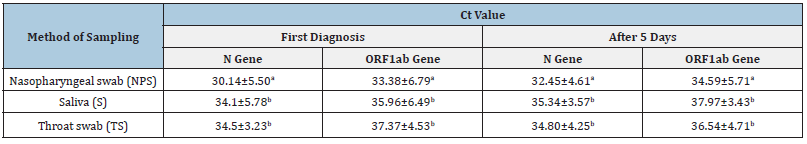
Values indicate means ± S.D.
Means ± S.D. within a column lacking a common superscript differ at p≤0.05.
Although there was a significant difference between the Ct values of both used genes (i.e N and ORF1ab) in our study, all tested samples were positive for COVID-19 and continued until 5 days after the first diagnosis. The virus load is significantly higher in NPS samples. NPS are the most accepted standard method for sample collection but are limited by their need for collection materials and well-trained healthcare professionals for appropriate sampling [13]. Our findings indicate the possible use of saliva as an alternative method to detect the virus in people who may suffer from health problems that prevent taking a nasopharyngeal swab. Saliva performs comparably to NPS for the detection of SARS-CoV-2 [13]. Saliva is simple for collection, can be used immediately, and can be tested with equipment readily available at most laboratories. Pasomsub et al. [14] found two cases had detectable SARS-CoV-2 from saliva samples, but not from nasopharyngeal and throat swabs.
Of these two saliva samples, the Ct values of the ORF1ab and N genes were 33.9 and 34.8, respectively, in one specimen, and 36.2 and 33.7, respectively, in another specimen. These two individuals later reported having anosmia which is an indicator for infection. Pojul et al. [10] found that N gene require longer duration of days with 12.68 (S.D.±3.24) to become negative than ORF1ab with 12.09 (S.D.±2.88) days and it differs significantly (p=0.012; p<0.05). Banko et al. [2] found that the positivity rate of saliva samples (81%) was higher than nasopharyngeal samples (71%). In contrast, González Losada et al. [5] reported results in which the virus was detected in 90%, 70% and 30% in samples collected from nose, throat, and saliva, respectively in north America. Ertuğrul et al. [3] confirmed that saliva samples can be used instead of nasopharyngeal samples in the detection of COVID-19, which is easier and less expensive. In France, Sandrine et al. [6] showed higher rate in nasopharyngeal swabs (5.81%) as compared to saliva swabs (0.39%) where the positive saliva sample was associated with a positive nasopharyngeal sample and no positive saliva sample was obtained in the absence of a positive nasopharyngeal sample. In China, Xiong et al. [4] showed that the highest rates were in the nasopharyngeal swab.
We found that 5 days after first diagnosis are not enough for the case to become negative for both N and ORF1ab genes regardless of the sampling method. RNA of SARS-CoV-2 was detected in saliva specimen after 37 days post onset in a man aged 71 years [15]. We also observed one positive case after 45 days of the first diagnosis by rRT-PCR with presence of high titer antibodies (unpublished). The reason could be that DNA copies of SARS-CoV-2 sequences might be integrated into the genome of the viral targeted cells. The integration and transcription of viral sequences may thus contribute to the detection of viral RNA by PCR in patients after infection and clinical recovery [16].
Conclusion
In conclusion, our results showed that the viral load was higher in NPS samples at first time of diagnosis and even after 5 days of infection. Saliva could be used as an alternative to NPS in people who suffer from health problems that prevent taking a nasopharyngeal swab. The time of sampling, disease status and clinical severity have to be taken into consideration. More research is required to determine the relationship between cell types of the buccal cavity and expression of housekeeping genes.
Acknowledgment
The authors thank all the diagnostic team at Biotechnology Research Center, Alzintan Branch for their valuable assistance. This work is a part of graduation project for B.Sc. students in the Department of Microbiology, Faculty of Science, Alzintan University.
References
- Ciotti M, Ciccozzi M, Terrinoni A, Jiang WC, Wang CB, et al. (2020) The covid-19 pandemic. Critical Reviews in Clinical Laboratory Sciences 57(6): 388-365.
- Banko A, Petrovic G, Miljanovic D, Loncar A, Vukcevic M, et al. (2021) Comparison and sensitivity evaluation of three different commercial real-time quantitative pcr kits for SARS-CoV-2 detection. Viruses 13(7): 1321.
- Ertuğrul G, Koroglu M, Yürümez Y, Toptan H, Kose E, et al. (2020) Comparison of saliva and oro-nasopharyngeal swab sample in the molecular diagnosis of COVID-19. Rev Assoc Med Bras 66(8): 1116-1121.
- Xiong W, Li T, Xu W, Weiyong L, Yanjun L, et al. (2020) Comparison of nasopharyngeal and oropharyngeal swabs for SARS-CoV-2 detection in 353 patients received tests with both specimens simultaneously. International Journal of Infectious Diseases 94: 107-109.
- González Losada C, González Lodeiro LG, Beato Canfux AI, Raúl Fernández J, Camacho H, et al. (2021) Comparison between nasopharyngeal swabs and saliva as reliable specimens for the diagnosis of SARS-CoV-2. Revista Habanera de Ciencias Médicas 20(3): e3745.
- Sandrine C, Catherine F, Baptiste D, Aurelien A, Jean Philippe L, et al. (2021) Comparison of nasopharyngeal and saliva swabs for the detection of RNA SARS-CoV-2 during mass screening (SALICOV study). New Microbiology 44(1): 59-61.
- Kammon A, Giweli A, Erhouma E, Rammah E, Elusta A, et al. (2021) Epidemiology of SARS-CoV-2 and emergence of UK variant in Zintan City of Libya. Open Journal of Epidemiology 11(4): 349-359.
- Corman VM, Drosten C (2020) SARS-CoV-2 detection by real-time RT-PCR. Eurosurveillance 25: 2001035.
- Rangaiah A, Shankar SM, Basawarajappa SG, Shah PA, Chandrashekar A, et al. (2021) Detection of SARS-CoV-2 in clinical samples: Target-specific analysis of qualitative Reverse Transcription-Polymerase Chain Reaction (RT-PCR) diagnostic kits. IJID Regions 1: 163-169.
- Pojul L, Vaishali S, Suranjana CH, Monjuri K, Dina R, et al. (2020) Dynamics of ORF1ab and N Gene among hospitalized COVID-19 positive cohorts: A hospital based retrospective study. medRxiv 11: 20236240.
- Skolimowska K, Rayment M, Jones R, Madona P, Moore LSP, et al. (2020) Non-invasive saliva specimens for the diagnosis of COVID-19: caution in mild outpatient cohorts with low prevalence. Clinical Microbiology and Infection 26(12): 1711-1713.
- Theda C, Hwang SH, Czajko A, Loke YJ, Leong P, et al. (2018) Quantitation of the cellular content of saliva and buccal swab samples. Scientific Reports 8: 6944.
- Kandel C, Zheng J, McCready J, Serbanescu MA, Racher H, et al. (2020) Detection of SARS-CoV-2 from saliva as compared to nasopharyngeal swabs in outpatients. Viruses 12(11): 1314.
- Pasomsub E, Watcharananan SP, Boonyawat K, Janchompoo P, Wongtabtim G, et al. (2021) Saliva sample as a non-invasive specimen for the diagnosis of coronavirus disease 2019: a cross-sectional study. Clinical Microbiology and Infection 27(2): 285.e1-285.e4.
- Tajima Y, Suda Y, Yano K (2020) A case report of SARS-CoV-2 confirmed in saliva specimens up to 37 days after onset: Proposal of saliva specimens for COVID-19 diagnosis and virus monitoring. Journal of Infection and Chemotherapy 26(10): 1086-1089.
- Zhang L, Richards A, Barrasa MI, Hughes SH, Young RA, et al. (2021) Reverse-transcribed SARS-CoV-2 RNA can integrate into the genome of cultured human cells and can be expressed in patient-derived tissues. Proceedings of the National Academy of Sciences 118(21): e2105968118.
© 2022 Abdulwahab Kammon. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






