- Submissions

Full Text
Advancements in Case Studies
Quantum microRNA Assessment of COVID-19 RNA Vaccine: Hidden Potency of BNT162b2 SASR-CoV-2 Spike RNA as MicroRNA Vaccine
Fujii YR*
Kawada-Cho, 106-6, Atsuta-Ku, Nagoya, Japan
*Corresponding author: Yoichi Robertus Fujii, Kawada-Cho, 106-6, Atsuta-Ku, Nagoya, Japan
Submission: March 05, 2021;Published: March 16, 2021

ISSN 2639-0531Volume3 Issue1
Abstract
Objective: The pandemic of coronavirus disease 2019 (COVID-19) is caused by infection with severe respiratory syndrome human coronavirus 2 (SARS-CoV-2). The spike (S) RNA of SARS-CoV-2 is used to build the COVID-19 vaccine. Although the level of immune response elicited by the full-length S RNA vaccine and the Receptor Binding Domain (RBD) vaccine that is a part of S is similar, the full-length S RNA vaccine is safer and lower reactogenicity than the RBD vaccine. However, the reason has not yet been clarified. On the other hand, the COVID-19 RNA vaccine with the S sequences may produce viral microRNAs (miRNAs). But there are no miRNA assessments for the safety of the COVID-19 vaccines. Therefore, evaluation as miRNA vaccines is necessary for risk management of vaccination.
Materials and Methods: miRNA Fold was used for pre-miRNA prediction in SARS-CoV-2 S RNA. MRmicro- T was used for protein target search. The integrated network algorithm as the miRNA entangling sorter (METS) analysis by using quantum miRNA language plus Artificial Intelligence (AI) were used as previously described.
Results: In computation, sixteen SARS-CoV-2-S-derived miRNAs bound to the negative strand S RNA with quite strong avidities. Further, CovS-miR-21 downregulated Rho associated coiled-coil containing protein kinase (ROCK2) and aryl hydrocarbon receptor nuclear translocator like (ARNTL), and CovSmiR- 3 decreased lysine methyltransferase 2C (KMT2C). Therefore, in the METS analysis, CovS-miR-21 suppressed the function of Ras homolog family member A (RhoA)/Rock2 signaling and circadian rhythm, and CovS-miR-3 inhibited histone H3-K4 methylation.
Conclusion: We found that BNT162b2 inhibits SARS-CoV-2 replication through degradation of negative strand viral RNAs that are completely paired with SARS-CoV-2 S-derived miRNAs. Further, CovS-miR-21 derived from BNT162b2 restores circadian rhythm and attenuate immunogenicity. Quantum miRNA assessments showed that the BNT162b2 RNA vaccine has a character of miRNA vaccine and is an excellent vaccine with high efficacy and low side-effects.
Keywords: COVID-19; SARS-CoV-2; BNT162b2; RNA vaccine; microRNA; Computer simulation; In silico; Circadian rhythm; Rho pathway
Introduction
In December 2019, severe respiratory syndrome human coronavirus 2 (SARS-CoV-2)
was identified in Wuhan, China [1]. This zoonotic RNA virus causes coronavirus disease 2019
(COVID-19) (World Health Organization, WHO) (www.who.int). The genome of SARS-CoV-2 is
a positive (+) single-stranded RNA with a nucleic acid length of approximately 30kd. Genome
analysis showed that SARS-CoV-2 belongs to the Coronaviridae family of the Beta-coronavirus
genus. The viral genome encodes three viral proteins within the envelope: the membrane
protein (M), the envelope protein (E), and the spike protein (S). The S protein is a transmembrane
protein that binds to host angiotensin-converting enzyme 2 (ACE2) for viral entry into
host cells.
The SARS-CoV-2 life cycle does not require nuclear transport. Virus replication requires
two different processes. One is the copy of the genome and the other is the production of
viral structural and accessory proteins from the sub-genomic mRNAs. The viral replication
hub protein, viral RNA-dependent RNA polymerase (Rd RP), is first translated by the host cell
mechanism from the infectious viral RNA orf1a/b. Rd RP then initiates negative strand RNA
(-) synthesis as complementary viral genomic RNA (+) (Figure 1) [2]. Its RNA (-) is involved in
two different processes of viral replication as a template.
Figure 1: Scheme of quantum miRNA assessments for RNA vaccine BNT162b2. BNT162b2 contains SARSCoV- 2 S mRNA. The S mRNA is sandwiched between 5’-UTR of human α-globin including the Kozak sequence and 3’-UTR of human ribosomal RNA. Further, 5’ terminal has cap formation and 3’ terminal has poly A tail. After incorporated into cells, mutated S protein (less viral function) is effectively induced by vaccine recipient’s translation system. Then, a part of S protein would be presented on the antigen-presenting macrophages. The neutral Abs could rise and Th1 would be promoted in vaccinated peoples. On the contrary, the S RNA would produce miRNAs and the miRNA may react to the negative strand SARS-CoV-2 RNA and target proteins. Quantum miRNA assessments should be necessary for understanding low incidents and the high efficacy of BNT612b2.
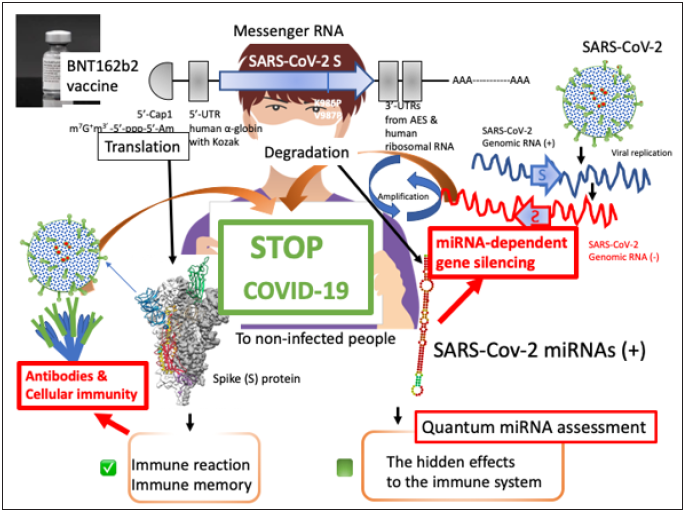
On the other hand, the reaction between the S protein and ACE2
is required to complete invasion of host cells, such as lung type II
alveolar cells. The Receptor-Binding Domain (RBD: amino acid
residue (aa)319-541) of the S protein, which is part of the N-terminal
S1 fragment, has been identified by crystal structural analysis [3].
After binding, the S protein is rearranged by membrane fusion with
the endosomal membrane via cleavage by the host protease. The
C-terminal furin (protease) cleavage fragment (S2) of the S protein
is deeply implicated in this fusion machinery. Stabilized mutations
of S2 from aa986 (lysine, K) and aa987 (valine, V) to proline (P)
have been reported to block the fusion state [4]. The S protein also
contains the antigenic determinants of neutralizing antibodies
(Abs) in COVID-19 patients [5]. Therefore, the S protein synthesized
from S RNA is a crucial factor in the development of SARS-CoV-2
vaccines to prevent the COVID-19 pandemic.
For RNA vaccines and viral vector-dependent DNA vaccines,
there are COVID-19 vaccine candidates using the S sequence [6].
BioNTech and Pfizer have developed two RNA vaccine candidates,
BNT162b1 and BNT162b2. BNT162b1 and BNT162b2 contain RBD
messenger RNA (mRNA) and total S protein mRNA, respectively
[7,8]. BNT162b1 and BNT162b2 elicited high titers of neutralizing
Abs in the sera of vaccinated individuals [9,10]. And all the data on
RNA vaccines are publicly available. Both vaccines are N1-methylpseudouridine
(Ψ) nucleoside-modified mRNA vaccine formulated
with Lipid-Nanoparticles (LNPs) and BNT162b2-derived S protein
contains stable mutations in K986P and V987P. Unmodified RNA
is highly immunogenic in mouse experiments, but N1-methyl-Ψ
RNA weakens innate immunity activities, such as interferon-α
production or activation of Toll-Like Receptor 3 (TLR3), and
modification of N1-methyl-Ψ mRNA increases its mRNA translation
[11,12].
The Ψ has been discovered in mRNA, ribosomal RNA (rRNA),
transfer RNA (tRNA) and small nuclear RNA in humans [13]. EMG1
N1-specific pseudouridine methyltransferase (EMG1, NEP1) can act to biosynthesize the N1-methyl-Ψ in eukaryotic 18S rRNAs
[14]. Therefore, the Ψ and N1-methyl-Ψ are naturally occurring
nucleotides. Additionally, when the Ψ was used for RNA interference
experiments, the Ψ containing small RNAs have served as an
efficient gene expression inhibitor [15]. Further, LNPs are composed
of four lipids, ((4-hydroxybutyl) azanediyl) bis(hexane-6,1-diyl)
bis(2-hexyldecanoate) (ALC-0315), the N, N-di-myristylamide of
2-hydroxyacetic acid (ALC-0159), 1,2-distearoyl-sn-glycero-3-
phosphocoline (DSPC) and cholesterol. The apparent terminal t1/2
in both the plasma and the liver were 2-3 and 6-8 days for ALC-
0159 and ALC-0315, respectively (www.gov.uk in Summary of the
Public Assessment Report for Pfizer/BioNTech COVID-19 vaccine).
Therefore, a rare allergic reaction or anaphylaxis by LNPs may be
conceivable just after injection of RNA vaccine solution but the
long-term effect of vaccine LNPs to our health might be less.
When comparing the safety and immunogenicity of the
BNT162b1 and BNT162b2 vaccines into young people (18-55 year
of age) and elderly people (65-85 year of age), BNT162b2 has been
lower incidences and lower severity of systemic side reactions than
BNT162b1 [10]. The immune response was equally elicited by the
RBD vaccine BNT162b1 and the total S RNA vaccine BNT162b2.
In addition, both vaccines share the same modified RNA platform,
the same RNA production and purification processes, and the same
LNP composition. Therefore, it is suggested that the RNA sequences
in the vaccine may influence the side-effect profile. Further, 15-28
days after the first dose of BNT162b2 vaccination, efficacy has been
recorded at 85-91%, despite of quite low titers of neutralizing Abs
[10,16]. However, this high efficacy of BNT162b2 has not yet been
elucidated, and the reason for the lower incidence of side reactions
with BNT162b2 vaccine remains unclear.
It has already been found that coronavirus-derived viral
miRNAs are encoded in the middle east respiratory syndrome
(MARS)-CoV genome [17], and SARS-CoV produced viral miRNAs
from the NSP3 region and N regions (svRNA-N). Anti-svRNA-N viral
miRNA reduced lung pathogenic effects in vivo [18]. For SARSCoV-
2, viral miRNAs have been identified in silico [19-21]. Since
viral miRNAs are generated from SARS-CoV genomic RNA, the
initial protective effect against SARS-CoV-2 on RNA vaccines during
the eclipse period until neutralization Abs rises is due to SARS-CoVderived
viral miRNAs. It is the miRNA-dependent gene silencing by
viral miRNAs derived from SARS-CoV-2 S RNA because SARS-CoV-2
S-derived miRNAs are perfectly paired with the negative strand S
RNA (-) of the viral genome. In fact, small RNAs that are completely
paired to the viral genomic RNAs inhibit the replication of influenza
and foot-and-mouth disease viruses by de-gradating genomic RNA
[22,23]. Additionally, Aydemir et al. [24] have computationally
predicted SARS-CoV-2 encoded miRNAs targets and the viral
miRNAs were related with gene silencing of nuclear factor kappa
B (NFKB), Janus kinase (JAK)/signal transducer and activator of
transcription (STAT) and transforming growth factor beta (TGFB);
however, SARS-CoV-2 S-derived miRNAs have not been listed in the
report. Thus, it will be considered that viral miRNAs from SARSCoV-
2 may have a hidden potency of reducing side-effect of the RNA
vaccine and may contribute to the initial antiviral defense response
of the RNA vaccine.
We have previously found by computing simulation that the
defense mechanism against viral infections, ‘the quantum miRNA
immunity’ in host is programmed by the quantum miRNA language
but viruses produce viral miRNAs to evade the miRNA immunity
[20,21,25]. This new discovery of host/virus interaction was
achieved by a bioinformatic analysis using miRNA Entangling
Target Sorter (METS) algorithm with the quantum miRNA Language
Plus Artificial Intelligence (MIRAI). Complements C1QA and
C1QB, Acetylcholine Esterase (ACHE) and Mitochondrial Antiviral
Signaling protein (MAVS)/Virus-Induced Signaling Adaptor (VISA)
have been targeted by a viral miRNA, Cov-miR-2 from the orf10
of the SARS-CoV-2 genome [20,21]. Suppression of complements,
ACHE and MAVS/VISA expression was implicated in weak response
and SLE-like symptom, augmenting inflammation of lung and
autonomic imbalance, and immune deficiency, respectively. It
showed that SARS-CoV-2 viral miRNA would cause pathogenicity of
COVID-19, in turn, the viral miRNA reduced host immunoreaction to
evade from the host defense system. It strongly suggests that SARSCoV-
2 RNA vaccine containing viral miRNAs would have hidden
potency to reduce the reactogenicity of RNA vaccine and block viral
replication via RNA-dependent gene silencing by breaking down
negative strand viral RNA (-). In this report, we investigated viral
pre-miRNA from SARS-CoV-2 S and the etiology of viral miRNAs
from the pre-miRNAs were simulated by METS with MIRAI.
Materials and Methods
Databases and prediction of SARS-CoV-2 S pre-miRNAs
Google Scholar (https://scholar.google.co.jp) was used for extraction of vaccine data. PubMed (www.ncbi.nlm.gov/pubmed/) were also used as a control database not to affect for AI analysis as a false premise bias of our papers listed. Total information content was 401,597 in vaccine, 298,180 in immunity and 42,548 in side-effects. The gene function of protein was searched by Gene- Cards (www.genecards.org). Protein ontology was investigated by GO enrichment analysis in Gene-ontology (geneontology.org). Prediction of pre-miRNAs in SARS-CoV-2 S sequences was computed online by miRNA-Fold (evryrna.ibisc.univ-evry.fr/miRNA Fold) according to human pre-miRNA structures. Viral miRNAs and their target information were searched with VIRmiRNA (crdd.osdd.net/ servers/virmirna/index.html). MR-microT in DIANA TOOLS (diana. imis.athena) was also used for direct protein target search. Data mining about viral miRNA was performed by functional analogy assay as described below.
METS analysis for BNT162b2 miRNAs
METS analysis was performed using SARS-CoV-2 S-derived miRNAs by the computer processing as described previously [26,27]. The quantum energy of miRNA was calculated by double miRNAs’ quantum energy levels of Double Nexus Score (DNS) entangling single miRNA quantum energy levels of Single Nexus Score (SNS). Data of multi-targets to a miRNA and a target to multimiRNAs was extracted from Target Scan Human 7.2 (targetscan.org) and miRTarBase Ver. 8.0 (mirtarbase.cuhk.edu.cn). Target protein/ protein interaction and protein cluster analysis were searched by STRING Ver. 11.0 (string-db.org). By using the quantum miRNA language plus artificial intelligence (MIRAI), the Area Under the Curve (AUC) in Receiver Operating Characteristic (ROC), accuracy, precision and F values were calculated as previous description [28,29].
Functional analogy assay and homology analysis
Data of viral miRNA and viral genome was obtained from miRBase Ver. 22.1 (miRbase.org) and Viral Genomes (ncbi.nim. nih.gov). SARS-CoV-2 S sequence (NC_045512.2) was used for calculation of binding ability with putative SARS-CoV-2 S-derived miRNAs. The functional analogy assay between viral miRNA and human miRNA was performed as previously described [29]. BLASTN in miRBase was used for homology sequence search of miRNAs. The RNA secondary structure and the binding ability measuring of RNA were computed by RNAFold (rna.tbi.univie. ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi).
Results and Discussion
Putative SARS-CoV-2 miRNAs from the S region
Pre-miRNAs were predicted from SARS-CoV-2 S RNA sequences using human (Homo sapiens) file parameters with the verified features of 70%. As shown in Figure 2, 74 pre-miRNA candidates were found, and the secondary structures of the pre-miRNAs were computed. The free energy of the stem-loop was calculated in miRNAFold, and pre-miRNA candidates with low levels of free energy (> -30 kcal/mol) were cut off. The RBD did not have pre-miRNA stem-loop sequences with high levels of free energy (Figure 2). Thirteen pre-miRNAs were selected and the secondary structure was computed in RNA fold server again. The putative 16 miRNA (CovS-miRNAs) in 11 pre-miRNAs were found from the homologous analysis to human miRNA sequences (Table 1). In functional homology assay, the 8 seed sequences of CovS-miR-21, CovS-miR-50.3, CovS-miR-55 and CovS-miR-60 were 88, 100, 88 and 88% homologous to hsa-miR-142-3p, hsa-miR-4793-3p, hsamiR- 1265 and hsa-miR-6881-3p, respectively (Table 1).
Figure 2: Prediction of pre-miRNA from BNT162b2. Using miRNA Fold web browser, pre-miRNA stem-loops (74 stems) in SARS-CoV-2 S RNA were extracted. Thirteen putative SARS-CoV-2 S-derived pre-miRNAs in Table 1 were depicted as the minimal free energy (ΔG, kcal/mol) (squared). The RBD region is shown as red arrows. Data from mRNA Fold is compared with that from RNA Fold server.
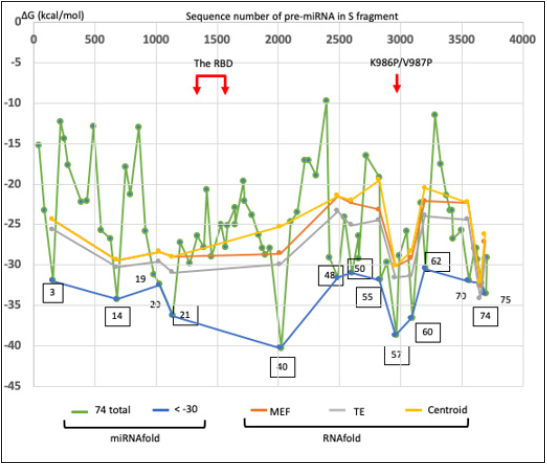
Table 1:Homologous sequences between SARS-CoV-2 S pre-miRNA and human miRNAs
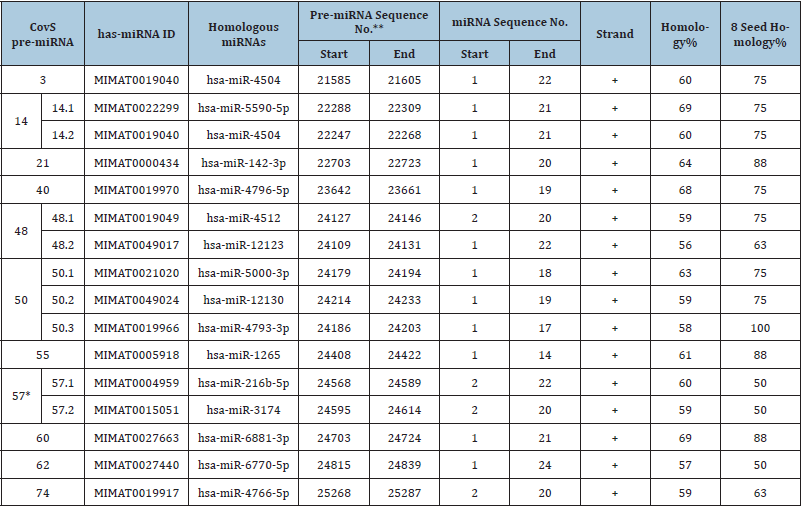
*The sites of K986P and V987P mutations in BNT162b2 S are contained in the pre-miRNA CovS-miR-57. **The region of RBD (aa319-541) is contained in pre-miRNA regions as shown in Figure 2.
Further, in MR-mciroT analysis, direct targets in Homo sapiens against the 8 seed of 16 miRNA candidates were extracted with the cut-off score of 0.99837, and 12 miRNAs were selected (data not shown). Target protein numbers of CovS-miR-3, CovS-miR-14, CovS-miR-21, CovS-miR-48.1, CovS-miR-48.2, CovS-miR-50.1, CovS-miR-50.2, CovS-miR-50.3, CovS-miR-55, CovS-miR-57.2, CovS-miR-60 and CovS-miR-62 were 9, 23, 2, 12, 1, 1, 1, 1, 4, 3, 4 and 16, respectively (data not shown).
Anti-SARS-CoV-2 avidity of BNT162b2-derived miRNAs
The CovS-miRNAs binding avidity was computed to negative and positive strand S targets (Table 2). Although the Ψ can be strongly paired to adenine rather than the pairing of intact uridine to adenine [13], the Ψ activities on RNA-dependent gene silencing are irregularly changed by both the position of Ψ and a Ψ content [15]. Therefore, we used uridine as a naïve molecule for calculation of the minimal free energy (ΔG, kcal/mol).
Table 2:The putative pre-miRNAs from SARS-CoV-2 S.
*kcal/mol
Sixteen CovS-miRNAs showed ultimate high levels of free energy to negative strand S targets compared with to positive strand S RNA, and the mean potential of whole affinity for negative strand S targets was -43.7 + 6.23 (mean + SD). Since Honeysuckle MIR2911 (5’ggccgggggacggacuggga3’) has been shown anti-SRASCoV- 2 activity in patients with -19.3 + 2.07 of ΔG [30-32], all CovSmiRNAs could inhibit replication of SARS-CoV-2. As 13.2μg/ml of MIR2911 completely inhibited virus replication in vitro, and 0.42- 1.13μg/ml of MIR2911 in serum exosome inhibited SARS-CoV-2 replication in patients, 10-30μg of BNT162b2 in vaccination can be enough source of anti-SARS-CoV-2 CovS-miRNAs to prevent viral replication, relevantly. Further, the Ψ can be strongly paired to adenine as described above, therefore, potency of the N1-methyl-Ψ containing CovS-miRNA molecules to anti-SARS-CoV-2 may be much higher than naïve CovS-miRNA ones. Thus, it is suggested that BNT162b2-derived miRNAs would play an important role for the early high efficacy (15-28 days after the first dose) in BNT162b2 vaccination to prevent viral infection (85-91%) despite of quite low titer upon neutralizing Ab production.
METS analysis for BNT162b2-derived miRNAs
METS analysis was performed to predict BNT162b2-derived miRNA functions by the mixed data from functional homology analysis and MR-mciroT analysis. As summarized on Table 3, targets were extracted, such as KMT2C to CovS-miR-3, NF1 to CovSmiR- 14, ARNL, RAC1, ROCK2, HMGA1 and MYB to CovS-miR-21, LRP-6, NFIB, PTPRD to CovS-miR-48.1, MBD3L1 to CovS-miR-50.3, MTRNR2L3, CYP7B1, GBP3, KRTAP2-3 and PEG3 to CovS-miR-55, and NR2F2 to CovS-miR-62. Four protein clusters, Rho signaling transduction (GO: 000726), WNT-activated receptor activity (GO: 0042813), histone H3-K4 methylation (GO: 0051568) and circadian rhythm (GO: 0007623) were obtained from protein/protein interaction and GO enrichment data (Figure 3 & 4). MBD3L1, MTRNR2L3, CYP7B1, GBP3 and KRTAP2-3 were not linked with these four clusters in protein/protein interaction analysis (data not shown). Therefore, the relation among 12 target proteins and BNT162b2-derived miRNAs was further investigated in the METSMIRAI analysis as quantum miRNA assessments.
Figure 3: MiRNA prediction from BNT162b2-derived pre-miRNAs. The putative miRNA (16 CovS-miRNAs) was found from the homologous analysis to human miRNA sequences (Table 2). CovS-miR-3 from SARS-CoV-2-PremiR- 3, CovS-miR-21 from SARS-CoV-2-Pre-miR-21, CovS-miR-57.1 and CovS-miR-57.2 from SARS-CoV-2-PremiR- 57 were depicted. The secondary structures of CovS-miR-57.1 and CovS-miR-57.2 (right square) were also shown in the small environment.
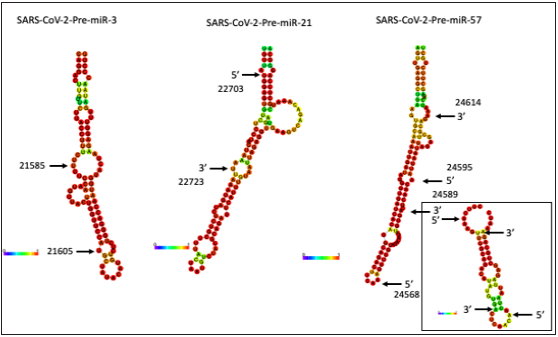
Table 3: METS analysis of CovS-miRNAs.
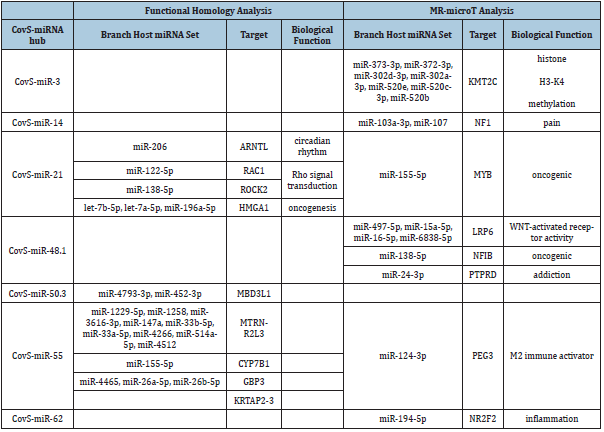
Figure 4:The network of METS simulation for CovS-miRNAs. The linkage among protein clusters and miRNAs in Table 3 was depicted by METS. miRNAs in red: upregulation, target proteins in blue squares: downregulation. Small circles in red are the hub miRNAs. Large circles show GO functions, circadian rhythm (GO: 0007623), WNT-activated receptor activity (GO: 0042813) and Rho signal transduction (GO: 0007266). Large trapezium is histone H3-K4 methylation (GO: 0051568).
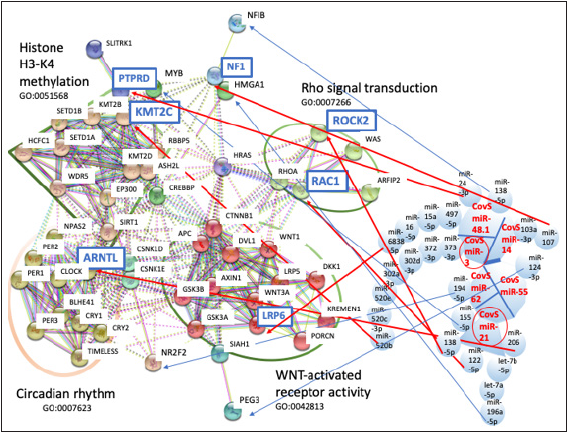
Anti-inflammation by CovS-miR-21
It is well known that a Rho-kinase, Rho associated coiledcoil
containing protein kinase (ROCK) is a pivotal effector of
the small GTP-binding Rho protein, and Rho/ROCK pathway
is involved in diverse immune functions, such as apoptosis,
proliferation, migration and inflammation [33]. In mouse models,
aberrant activation of RhoA/ROCK2 pathway has been observed
in Rheumatoid Arthritis (RA), Systematic Lupus Erythematosus
(SLE) and Multiple Sclerosis (MS) via excessive production of
interleukin-17 (IL-17) and IL-21 [34]. Therefore, it is also known
that RhoA/ROCK2 pathway is related with T and B cell activation.
Further, RAC1 GTPase controls neutrophil recruitment into
inflamed and infected tissues via cytoskeletal dynamics [35]. CovSmiR-
21 inhibited ROCK2 and RAC1 with miR-138-5p and with miR-
122-5p (Figure 4). These results showed that CovS-miR-21 would
inhibit migration and activation of neutrophil, T and B cells, and
would attenuate inflammation through BNT162b2 intramuscular
(i.m.) injection in situ.
In addition, LDL receptor related protein 6 (LRP6) expression
was reduced by CovS-miR-48.1 with miR-497-5p, miR-15a-5p,
miR-16-5p and miR-6838-5p (Figure 4). Since LRP6 is a receptor
of proto-oncogene WNT and a secreted WNT (sWNT) agonist
Dickkopf-1 (DKK-1) has prevented LPS-induced inflammatory
responses in human bronchial epithelial cells via inhibition of
LRP6/sWNT [36], suppression of LRP6 by CovS-miR-48.1 would
also contribute for attenuation of inflammation through LRP6/
sWNT pathways. Nuclear receptor subfamily 2 group F member 2
(NR2F2, COUP-TFII) was suppressed by CovS-miR-62 with miR-194-
5p (Figure 4). NR2F2 has been involved into human endometrial
stromal inflammation in silico [37]; therefore, CovS-miR-62 may
also attenuate inflammation by BNT162b2 i.m. injection in situ.
Additionally, paternally expressed gene 3 (PEG3) were reduced
by CovS-miR-55 with miR-124-3p. As PEG3 expression has
been decreased in human glioma cells and tissues and reducing
PEG3 renders decreasing of CD8+ killer T cell proliferation and
suppression of IFNγ and TGF-β levels [38], downregulation of PEG3
may decrease M2 macrophage activity and ability of killer T cells.
Taken together, BNT162b2-derived miRNAs could be implicated in
reducing systemic events of vaccine recipients.
A balance of reactogenicity and immunogenicity by CovS-miR-3
Macrophage as an antigen-presenting dendritic cell is the center cell of the innate immunity. A key feature of macrophages is plasticity, and the process of macrophage plasticity induces two different phenotypes, M1 and M2. Classical M1 macrophage is induced by lipopolysaccharide (LPS) or interferon γ (IFNγ) and promotes a proinflammatory response via dominant Toll-like receptor (TLR) ligand or agonist. Alternative M2 macrophage is elicited from immune complex stimulation, and IL-4 or IL-13 production, which promote an anti-inflammatory response [39]. Histone H3 trimethylation on lysine 4 (H3K4me3) and 27 (H3K27me3) has been significantly correlated with macrophage differentiation [40]. In mouse model, M1 showed decreased global enrichment of H3K4me3 whereas M2 increased enrichment of H3K4me3 [41]. As CovS-miR-3 inhibited lysine methyltransferase 2C (KMT2C) with miR-373-3p, miR-372- 3p, miR-302d-3p, miR-302a-3p, miR-520e, miR-520c-3p and miR- 520b (Figure 4), H3K4me3 would be decreased by BNT162b2 vaccination, which may induce M1 state of macrophages. M1-like polarized macrophage promotes the Th1 polarization of CD4 cells [42], which has been confirmed upon BNT162b2 vaccination by Polack et al. [8] and the high efficacy of whole S BNT162b2 was documented [16]. It is suggested that CovS-miR-3 from BNT162b2 could successfully accelerate neutralization Ab production and Th1 cellular immunity response via M1 macrophage/Th1 polarization. Given anti-inflammation function by BNT162b2-derived CovSmiR- 21, a balance of reactogenicity and immunogenicity would be controlled by another BNT162b2-derived CovS-miR-3.
Pain-related CovS-miR-14
Neurofibromin 1 (NF1) gene on chromosome 17 in human is thought to be associated with neuro-fibromatosis type 1 and to be involved in pain [43]. Since CovS-miR-14 suppressed NF1 with miR-103a-3p and miR-107 (Figure 4), Covs-miR-14 may reduce pain after injection. Furthermore, receptor-type protein tyrosine phosphatase D (PTPRD) was suppressed by CovS-miR-48.1 with miR-24-3p (Figure 4). Decreasing of PTPRD has reduced cocaine reward and addiction in mice [44], so CovS-miR-48.1 may contribute for anti-addiction medication.
Anti-oncogenic CovS-miR-48.1 and CovS-miR-21
Nuclear Factor IB (NFIB) and v-myb avian Myelobastosis viral oncogene (MYB) were inhibited by CovS-miR-48.1 with miR-138- 5p and CovS-miR-21 with miR-155-5p, respectively (Figure 4). The MYB/NFIB fusion proto-oncogene generates the MYB/NFIB transcription factor and has been detected in a high percentage of patients with adenoid cystic carcinoma [45]. Further, High Mobility Group A1 (HMGA1) was suppressed by CovS-miR-21 with let- 7b-5p, let-7a-5p and miR-196a-5p (Figure 4). Since HMGA1 is an oncofoetal transcriptional factor and can be related with promotion and metastasis of numerous malignant cancers [46], both results are strongly suggested that BNT162b2 is non-oncogenic. Thus, oncogenic activity of BNT162b2 might be prevented by BNT162b2- derived miRNAs.
Circadian rhythm controller of CovS-miR-21
The robustness of circadian rhythm is implicated in the oscillating expression of clock gene family, such as clock circadian regulator (CLOCK), Aryl Hydrocarbon Receptor Nuclear Translocator Like (ARNTL, BMAL1), Period Circadian Regulators (PERs), Cryptochrome Circadian Regulators (CRYs). Disturbance of these clock genes lead to pathophysiology for cardiovascular system, whereas the stabilization is protective [47]. Although ARNTL was reduced by CovS-miR-21 with miR-206 (Figure 4), deletion of circadian rhythm-related gene BMAL1 has exacerbated acute bronchiolitis caused Sendai and influenza A virus in mice [48]. Therefore, CovS-miR-21 is an aggravating factor for SARS-CoV-2 pathogenicity because low levels of BMAL1 expression promoted RNA viral replication [49,50]. As shown in Figure 5, ARNTL expression is various among seasons in human blood sample and the lowest levels of ARNTL has been observed during the winter months [51] coinciding with the peak season for respiratory viral epidemics (Figure 5, a right panel) [52]. In the case of COVID-19 pandemic, since ARNTL was suppressed by Covs-miR-21 from SARS-CoV-2 S, infectivity and mortality of SARS-CoV-2 infection are acceleratively both higher in colder climates [53].
Figure 5: Summary of the quantum miRNA assessment for BNT162b2. BNT162b2-derived miRNAs inhibit SARS-CoV-2 replication via degradation of the negative strand viral RNA by completely paired RNA-dependent gene silencing (blue arrows), which would induce the high efficacy (85-91%) on 15-28 days after the first dose of BNT162b2 vaccination. BNT162b2-derived CovS-miR-21 would restore circadian rhythm and attenuate immunogenicity while CovS-miR-3 would induce Th1 promotion and antibody production. Therefore, CovSmiR- 21 and CovS-miR-3 would play an important role for lower incidence and high neutral Ab production plus Th1 promotion, respectively. The circadian oscillation is seasonally synchronized with immune cells’ gene expression in healthy people (the right panel) and SAR-CoV-2 S-derived CovS-miR-21 would make a narrow peak in the summer season (red arrows). On the other hand, BNT162b2-derived CovS-miR-21 could restore the circadian rhythm by direct beating the viral genome. Substantially as described as similia similibus curantur, BNT162b2-CovS-miR-21 would attenuate immunogenicity via suppression of clock gene. The pandemic of COVID-19 would be successfully decreased by BNT162b2.
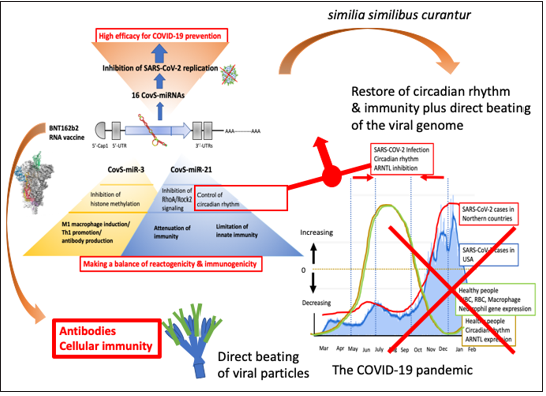
On the contrary, CovS-miR-21 from BNT162b2 would limit innate immunity via decreasing of ARNTL in circadian-rhythm [54,55]. ARNTL knockout mice has rendered macrophages unable to sustain inflammatory reaction and M1 macrophages enhanced ARNTL expression [56,57]. Thus, these data strongly suggest that inflammation by vaccine injection could be attenuated by CovSmiR- 21 as described above. Subsequently, since CoVS-miR-21 was a viral pathogenic factor, adenovirus- or vaccinia virus-based DNA vaccines should consider additional side-effects of SARS-CoV-2 S-derived miRNAs as circadian rhythm disturber because SARSCoV- 2 S DNA in adenovirus vector may be integrated into the host chromosome as a character of viral vectors [6,58,59]. For example, as the relationship of subarachnoid hemorrhage to circadian oscillation via CRYs and PER2 has been widely studied [47,60,61] spontaneous brain injury may be induced by SARS-CoV-2 S-derived miRNA.
The hub of miRNA/protein for BNT162b2 safety and efficacy
The results of the quantum miRNA assessment were validated by MIRAI about four subjects as vaccine, immunity, acute inflammation, side-effects (Table 4). An AUC of BNT162b2-derived miRNAs was 73.08% when data was compared with that of host miRNA upon SARS-CoV-2 infection (92.36%). Therefore, miRNA assessments with the METS analysis were statistically significant by AI validation. From high precision percentage (95.00%), it is suggested that CosV-miR-21/RhoA-Rock2 signaling/circadian rhythm axis and CovS-miR-3/histone H3-K4 methylation are the hub of BNT162b2 safety and efficacy, respectively (Figure 5).
Table 4: The validation of miRNA assessment upon RNA v.

*precentage
Given SARS-CoV-2 S-derived miRNAs, with respect to the SARSCoV-
2 pandemic, ARNTL expression in low level season increase
COVID-19 infection cases, in contrast, ARNTL in high level season
decrease COVID-19 according to circadian rhythm. In viral infection,
SARS-CoV-2 would escape from quick immune response by
effective macrophage-phenotype induction in host by using RhoA/
Rock2 inhibition and would limit immune reaction by controlling
of circadian rhythm. Therefore, BNT162b2 vaccine-derived CovSmiRNAs
would work as ‘similia similibus curantur ‘. Although
accuracy and F value were not statistically enough in the validation
of BNT162b2, the low percentages of accuracy (48.70%) and F
value (64.41%) might be due to biphasic characters of BNT162b2
S as this time limitation of the AI-validation. Because the control
host miRNAs in SARS-CoV-2 infection showed high percentage of
accuracy (90.00%) and F values (94.34%) (Table 4). Thus, more
data may be necessary for assessment of safety and efficacy on
COVID-19 RNA vaccine.
We have previously shown that rice MIR2097-5p putative
binding site was broadly distributed in the viral genome of SARSCoV-
2 [32]. However, CovS-miRNAs did not show significant
binding avidity to the pathogenic orf10 of SARS-CoV-2. It suggests
that RNA vaccination may not be enough to completely silence the
COVID-19 pandemic. Since our study shows that an approach to
design a mix of miRNAs targeting several regions of the SARS-CoV-2
open reading frame (orf) and the UTRs would be possible in the
development of miRNA vaccines, plant MIR2079-5p mixed miRNA
vaccine may be applicable for advanced BNT162b2 vaccines [62].
In addition, patients who developed COVID-19 after vaccination
may need therapeutic agents such as rice MIR2097-5p to control
SARS-CoV-2. Further investigations are needed.
Conclusion
On the premise of conclusion, in BNT162b2 vaccine, LNPs is used as a non-viral delivery system, contributing to vaccine safety and an effective immune response. Our study in silico highlights the importance of BNT162b2 vaccine-derived miRNAs as a hallmark of miRNA vaccines and reveals a novel molecular mechanism of RNA vaccine efficacy through an excellent balance of reactogenicity and immunogenicity. We found that BNT162b2-derived CovSmiR- 21 and its target ARNTL and ROCK2 emerge as key player in the restoration of circadian rhythms and the attenuation of immunogenicity, while a histone H3-K4 methyl transferase, KMT2C inhibition by CovS-miR-3 induces Th1 promotion and antibody production via M1 macrophage/Th1 polarization. The immune balance of this RNA vaccine significantly results higher efficacy and low side-effects as described as similia similibus curantur. Quantum miRNA assessments help validate vaccines to investigate hidden effects on the immune system.
References
- Zhu N, Zhang D, Wang W, Li X, Yang B, et al. (2020) A novel coronavirus from patients with pneumonia in China, 2019. New Engl J Med 382(8): 727-733.
- Romano M, Ruggiero A, Squeglia F, Maga G, Berisio R (2020) A structural view of SARS-CoV-2 RNA replication machinery: RNA synthesis, proofreading and final capping. Cells 9(5): 1267.
- Lan J, Ge J, Yu J, Shan S, Zhou H, et al. (2020) Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581(7807): 215-221.
- Kirchdoerfer RN, Wang N, Pallesen J, Wrapp D, Turner HL, et al. (2018) Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis. Sci Rep 8(1): 15701.
- Chi X, Yan R, Zhang J, Zhang G, Zhang Y, et al. (2020) A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science 369(6504): 650-655.
- Folegatti PM, Ewer KJ, Aley PK, Angus B, Becker S, et al. (2020) Safety and immunogenicity of the ChAdOx1nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomized controlled trial. Lancet 396(1024): 467-478.
- Mulligan MJ, Lyke KE, Kitchin N, Absalon J, Gurtman A, et al. (2020) Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature 586(7830): 589-593.
- Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, et al. (2020) Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. New Engl J Med 383(27): 2603-2615.
- Sahin U, Muik A, Derhovanessian E, Vogler I, Kranz LM, et al. (2020) COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell response. Nature 586(7830): 594-599.
- Walsh EE, Frenck R, Falsey AR, Kitchin N, Absalon J, et al. (2020) Safety and immunogenicity of two RNA-based Covid-19 vaccine candidates. New Engl J Med 383(25): 2439-2450.
- Karikó K, Muramatsu H, Welsh FA, Ludwig J, Kato H, et al. (2008) Incorporation of pseudo-uridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol Ther 16(11): 1833-1840.
- Andries O, Mc Cafferty S, De Smedt SC, Weiss R, Sanders NN, et al. (2015) N(1)-methyl-pseudo-uridine-incorporated mRNA outperforms pseudo-uridine-incorporated mRNA by providing enhanced protein expression and reduced immunogenicity in mammalian cell lines and mice. J Control Release 217: 337-344.
- Boo SH, Kim YK (2020) The emerging role of RNA modifications in the regulation of mRNA stability. Exp Mol Med 52(3): 400-408.
- Wurm JP, Meyer B, Bahr U, Held M, Frolow O, et al. (2010) The ribosome assembly factor Nep1 responsible for Bowen-Conradi syndrome is a pseudouridine-N1-specific methyltransferase. Nucleic Acids Res 38(7): 2387-2398.
- Sipa K, Sochacka E, Kazmierczak-Baranska J, Maszewska M, Janicka M, et al. (2007) Effect of base modifications on structure, thermodynamic stability, and gene silencing activity of short interfering RNA. RNA 13(8): 1301-1316.
- Amit S, Regev-Yochay G, Afek A, Kreiss Y, Leshem E (2020) Early rate reductions of SARS-CoV-2 infection and COVID-19 in BNT162b2 vaccine recipients. Lancet 397(10277): 875-877.
- Hasan M, Akter R, Ullah S, Abedin J, Ullah A, et al. (2014) A computational approach for predicting role of human microRNAs in MARS-CoV genome. Adv Bioinformatics 2014: 967946.
- Morales L, Oliveros JC, Fernandez-Deigado R, tenOever BR, Enjuanes L, et al. (2017) SARS-CoV-encoded small RNAs contribute to infection-associated lung pathology. Cell Host Microbe 21(3): 344-355.
- Khan MA, Sany MRU, Islam MS, Islam ABMMK (2020) Epigenetic regulator miRNA pattern differences among SARS-CoV, SARS-CoV-2, and SARS-CoV-2 World-Wide isolates delineated the mystery behind the epic pathogenicity and distinct clinical characteristics of pandemic COVID-19. Front Genet 11: 765.
- Fujii YR (2020) The etiology of COVID-19 in silico by SARS-Cov-2 infection with the quantum microrna language-AI. Virol Immunol J 4(2): 000243.
- Fujii YR (2020) The COVID-19 deadly risk assessment upon the updated etiologic computer simulation by quantum microRNA language in SARS-CoV-2 infection in eo. Int J Clin Case stud Rep 3(1): 142-154.
- Chen W, Yan W, Du Q, Fei L, Liu M, et al. (2004) RNA interference targeting VP1 inhibits foot-and-mouth disease virus replication in BHK-21 cells and sucking mice. J Virol 78(13): 6900-6907.
- Tompkins SM, Lo CY, Tumpey TM, Epstein SL (2004) Protection against lethal influenza virus challenge by RNA interference in vivo. Pro Natl Acad Sci USA 101(23): 8682-8686.
- Aydemir MN, Aydemir HB, Korkmaz EM, Budak M, Cekin N, et al. (2021) Computationally predicted SARS-CoV-2 encoded microRNAs target NFKB, JAK/STAT and TGFB signaling pathways. Gene Rep 22: 101012.
- Fujii YR (2020) The quantum microRNA immunity in human virus-associated diseases: virtual reality of HBV, HCV and HIV-1 infection, and hepatocellular carcinogenesis with AI machine learning. Arch Clin Biomed Res 4: 089-129.
- Fujii YR (2013) The RNA gene information: retroelement-microRNA entangling as the RNA quantum code. Methods Mol Biol 936: 47-67.
- Fujii YR (2018) Quantum language of microRNA: application for new cancer therapeutic targets. Methods Mol Biol 1733: 145-157.
- Fujii YR (2019) Cancer simulation from stage minus one by quantum microRNA language: lung, colorectal and pancreatic cancers. Med One 4: e190023.
- Fujii YR (2019) Quantum microRNA network analysis in gastric and esophageal cancers: xenotropic plant microRNAs cure from cancerous paradox via Helicobacter pylori infection. Gastroenterol. Hepatol Endosc 4: 1-18.
- Zhou Z, Zhou Y, Jiang XM, Wang Y, Chen X, et al. (2020) Decreased HD-MIR2911 absorption in human subjects with the SIDT1 polymorphism fails to inhibit SARS-CoV-2 replication. Cell Discovery 6: 63.
- Zhou LK, Zhou Z, Jiang XM, Zheng Y, Chen X, et al. (2020) Absorbed plant MIR2911 in honeysuckle decoction inhibits SARS-CoV-2 replication and accelerates the negative conversion of infected patients. Cell Discovery 6(1): 54.
- Fujii YR (2020) In silico study by quantum microRNA language for development of anti-COVID-19 agents: COVID-19 is prevented by rice MiR2097-5p through suppression of SARS-Cov-2 viral microRNAs plus HIPk2 target proteins. Virol Immunol J 4(4): 000256.
- Ricker E, Chowdhury L, Yi W, Pernis AB (2016) The RhoA-ROCK pathway in the regulation of T and B cell responses. F1000Res 5: F1000 Faculty Rev-2295.
- Biswas PS, Gupta S, Chang E, Song L, Stirzaker RA, et al. (2010) Phosphoryaltion of TRF4 by ROCK2 regulates IL-17 and IL-21 production and the development of autoimmunity in mouse. J Clin Invest 120(9): 3280-3295.
- Pantarelli C, Welch HC (2018) Rac-GTPase and Rac-GEFs in neutrophil adhesion, migration and recruitment. Eur J Clin Invest Suppl2(Suppl Suppl 2): e12939.
- Jang J, Jung Y, Kim Y, Jho EH, Yoon Y (2017) LPS-induced inflammatory response is suppressed by Wnt inhibitors, Dickkopf-1 and LGK974. Sci Rep 7: 41612.
- Li X, Large MJ, Creighton CJ, Lanz RB, Jeong JW, et al. (2013) COUP-TFII regulates human endometrial stromal genes involved in inflammation. Mol Endocrinol 27(12): 2041-2054.
- Yang F, Wang T, Du P, Fan H, Dong X, et al. (2020) M2 bone marrow-derived macrophage-derived exosomes shuffle microRNA-21 to accelerate immune escape of glioma by modulating PEG3. Cancer Cell Int 20: 93.
- Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, et al. (2014) Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity 41(1): 14-20.
- Tserel L, Kolde R, Rebane A, Kisand K, Org T, et al. (2010) Genome-wide promoter analysis of histone modifications in human monocyte-derived antigen presentating cells. BMC Genomics 11: 642.
- Ruenjaiman V, Butta P, Leu YW, Pongpanich M, Leelahavanichkul A, et al. (2020) Profile of histone H3 lysine 4 trimethylation and the effect of lipopolysaccharide/immune complex-activated macrophages on endotoxemia. Front Immunol 10: 2956.
- Atri C, Guerfali FZ, Laouini D (2018) Role of human macrophage polarization in inflammation during infectious disease. Int J Mol Sci 19(6): 1801.
- Bellampalll SS, Khanna R (2019) Towards a neurobiological understanding of pain in neurofibromatosis type 1 (NF1): mechanisms and implications for treatment. Pain 160(5): 1007-1018.
- Uhi GR, Martinez MJ, Paik P, Sulima A, Bi GH, et al. (2018) Cocaine reward is reduced by decreased expression of receptor-type protein tyrosine phosphatase D (PTPRD) and by a novel PTPRD antagonist. Proc Natl Acad Sci USA 115(45): 11597-11602.
- Mikse O, Tchaicha JH, Akbay EA, Chen L, Bronson RT, et al. (2016) The impact of the MYB-NFIB fusion proto-oncogene in vivo. Oncotarget 7(22): 31681-31688.
- Wang Y, Hu L, Zheng Y, Gao L (2018) HMGA1 in cancer: cancer classification by location. J Cell Mol Med 23(4): 2293-2302.
- Nogueira AB, Nogueira AB, Veiga JCE, Teixeira MJ (2017) Hypothesis on the role of cryptochromes in inflammation and subarachnoid hemorrhage outcome. Front Neurol 8: 637.
- Ehlers A, Xie W, Agapov E, Brown S, Steinberg D, et al. (2017) BMAL1 links the circadian clock to viral airway pathology and asthma phenotypes. Muc Immunol 11(1): 97-111.
- Edgar RS, Stangherlin A, Nagy AD, Nicol MP, Efstathiou S, et al. (2016) Cell autonomous regulation of herpes and influenza virus infection by the circadian clock. Proc Natl Acad Sci USA 113(36): 10085-10090.
- Majumdar T, Dhar J, Patel S, Kondratov R, Barik S (2017) Circadian transcription factor BMAL1 regulates innate immunity against select RNA viruses. Innate Immun 23(2): 147-154.
- Dowell SF (2001) Seasonal variation in host susceptibility and cycles of certain infectious diseases. Emerg Infect Dis 7(3): 369-374.
- Dopico XC, Evangelou M, Ferreira RC, Guo H, Pekalski ML, et al. (2015) Widespread seasonal gene expression reveals annual difference in human immunity and physiology. Nat Commun 6: 7000.
- Liu X, Huang J, Li C, Zhao Y, Wang D, et al. (2021) The role of seasonality in the spread of COVID-19 pandemic. Environ Res 195: 110874.
- Keller M, Mazuch J, Abraham U, Eom GD, Herzog ED, et al. (2009) A circadian clock in macrophages controls inflammatory immune responses. Proc Natl Acad Sci USA 106(50): 21407-21412.
- Zhuang X, Rambhatla SB, Lai AG, McKeating JA (2017) Interplay between circadian clock and viral infection. J Mol Med 95(12): 1283-1289.
- Alexander RK, Liou YH, Knudsen NH, Starost KA, Xu C, et al. (2021) Bmal1 integrates mitochondrial metabolism and macrophage activation. Elife 9: e54090.
- Early JO, Menon D, Wyse CA, Cervantes-Silva MP, Zaslona Z, et al. (2018) Circadian clock protein BMLA1 regulates IL-1β in macrophages via NRF2. Proc Natl Acad Sci USA 115(36): E8460-E8468.
- Stephen SL, Montini E, Sivanandam G, Al-Dhalimy M, Kestler HA, et al. (2010) Chromosomal integration of adenoviral vector DNA in vivo. J Virol 84(19): 9987-9994.
- Lehrman S (1999) Virus treatment questioned after gene therapy death. Nature 401(6753): 517-518.
- Frase S, Kaiser S, Steimer M, Selzner L, Foit NA, et al. (2021) Patients with subarachnoid hemorrhage exhibit disturbed expression of patterns of the circadian rhythm gene Period-2. Life 11(2): 124.
- Schallner N, Lieberum JL, Gallo D, LeBlanc III RH, Fuller PM, et al. (2017) Carbon monoxide preserves circadian rhythm to reduce the severity of subarachnoid hemorrhage in mice. Stroke 48(9): 2565-2573.
- El-Nabi SH, Elhiti M, El-Sheekh, M (2020) A new approach for COVID-19 treatment by microRNA. Med Hypotheses 143: 110203.
© 2020 Shi K. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






