- Submissions

Full Text
Modern Research in Dentistry
Reliability of Periodontal Pathogens and Human Cathelicidine Ll-37 in the Prediction of the Different Stages of Periodontal Disease
Zeyad Nazar Majeed1, Alabsi AM2, Dasan Swaminathan3 and Koshy Philip4*
1Department of Periodontology, Faculty of Dentistry, University of Babylon, Iraq
2Faculty of Dentistry, MAHSA University, Malaysia
3Department of Restorative Dentistry, Lincoln University College, Malaysia
4Lincoln University College, Petaling Jaya, Selangor, Malaysia
*Corresponding author:Koshy Philip, Lincoln University College, Petaling Jaya, Selangor, Malaysia
Submission: August 16, 2019;Published: September 03, 2019

ISSN:2637-7764Volume4 Issue3
Abstract
Objectives: The aim of this study was to determine the levels of LL-37 and subgingival periodontal pathogens in the periodontal health and disease among three ethnic groups in Malaysia.
Methods: Gingival crevicular fluid (GCF) and subgingival plaque samples from 28 healthy subjects (H), 30 patients with gingivitis (G) and 30 patients with chronic periodontitis (CP) were collected. The LL- 37 levels were quantified by the enzyme-linked immunosorbent assay (ELISA) while the subgingival periodontal pathogens were identified using polymerase chain reaction (PCR).
Result: The LL-37 levels were significantly higher in the CP group than other groups. The detectability of LL-37 in CP group was 100%, while in H and G groups was 78.5% and 93.3% respectively. Red complex species populations in CP group are higher than in other groups. The LL-37 levels in GCF were positively correlated with red complex and A. actinomycetemcomitans species in the subgingival plaque. Moreover, there is a positive correlation between the LL-37 levels and periodontal pathogens which play major role in the initiation and progression of chronic periodontitis.
Conclusion: LL-37 in GCF may be considered as relevant biomarker for chronic periodontitis. Furthermore, red complex species count can be useful in the differentiation between the different stages of periodontal disease.
Keywords: Human cathelicidine LL-37; GCF; Periodontal pathogens; Biomarkers
Introduction
Periodontal diseases are basically classified into two types that include gingivitis and chronic periodontitis. Both diseases are essentially caused by bacteria present in the subgingival biofilm. The determination of microbial profiles that could categorize various forms of periodontal disease has not been described [1]. The complex periodontal microbiota, in addition to its great variability among the individuals, may lead to these difficulties [2,3]. Furthermore, it has been proven that there are variances in the distribution of periodontal bacteria among different populations [4]. Along with these findings, the majority of these bacterial species that are also present in the normal periodontal microbiota will further complicate the determination of microbial profiles in subgingival plaque [5]. The existence of low-level periodontal pathogens in the periodontally healthy subjects is a common finding [6].These features of periodontal microbiota which can lead to inflammation justify the needs of the development of a more accurate microbial diagnosis which can support the clinical diagnosis and lead to a better treatment plan [7]. Five microbial complexes repetitively exist in the subgingival biofilm of individuals with and without periodontal inflammation [8]. This comprises the red complex which encompasses a conglomerate of three species namely Porphyromonas gingivalis, Tannerella forsythia and Treponema denticola.
The red complex has been considered as the most pathogenic bacterial complex [8,9] and may apply an inhibitory role in the colonization ability of other types of bacteria in the subgingival biofilm. Thus, they may have the ability to adjust the virulence characteristics of other subgingival biofilm communities [10]. Furthermore, Aggregatibacter actinomycetemcomitans (A.a) is considered as a causative pathogen for adult periodontitis and aggressive periodontitis. Moreover, it is suggested that A.a causes apoptosis to increase in the gingival epithelial cells [11,12]. Host defence antimicrobial peptides (AMPs) are the major components of the innate immune responses which have an essential role in human health [13]. More than 45 AMPs have been recognized in the oral cavity [14,15]. LL-37 has been distinguished as an exceptional class of peptides which has many effects including antibacterial, antiviral and antifungal functions. It also shows a chemotactic and modulatory effect and has the ability to induce wound healing, modifying apoptosis and angiogenesis. The antimicrobial action of the LL-37 occurs with other antimicrobial molecules such as histamine, defensins and enzymes that have the capacity to destroy the bacterial cell wall. Collectively, these molecules have the ability to respond rapidly and adequately to the bacterial challenges [16]. The terminology of mature cathelicidine LL-37 is derived from two leucine residues and 37 amino acids.
The mass of human cathelicidine antimicrobial peptide prior to the proteolytic processing was 18 KD and this is the reason why the term hCAP-18 has been used [17]. Therefore, the extracellular processing of hCAP-18 by proteinase 3 in response to phagocytosis or neutrophils stimulation will lead to the release of mature LL- 37 [18]. The LL-37 levels in saliva and GCF present less than the minimum inhibitory concentration for oral pathogens. This led to the suggestion that the main action of LL-37 and other AMPs is a trigger to the immune system, i.e. an endogenous mediator that enhances and recruit’s antigen-presenting cells to activate the innate and adaptive immunity [19]. Previous studies have demonstrated the expression of LL-37 in different stages of the periodontal disease progression [20,21] which determined the role of LL-37 in the innate immune system. We thus hypothesized that the LL-37 levels in GCF can be used as a biomarker for periodontal diseases. The aim of the present study was to correlate the LL-37 levels to the most common periodontal pathogens responsible for the initiation and progression of periodontal diseases. We also determined the correlation between the LL-37 levels and clinical periodontal parameters used to classify the periodontal disease and compared the levels of LL-37 between different Malaysian ethnic groups to investigate the effect of genetic variations on the LL-37 levels.
Material and Methods
Subjects
Patients attending the undergraduate and postgraduate Periodontal clinic, Faculty of Dentistry, University of Malaya were enrolled in this study. Subjects diagnosed as healthy, with generalized gingivitis and moderate to severe chronic periodontitis and those who agreed to participate were included in the study. The exclusion criteria included women who were pregnant and patients with systemic disease or infection, that have an effect on the immune system response, periodontal treatment in the past six months and antibiotics intake in the past three months. Smokers were also excluded from the study. Recruited subjects signed a consent form before their enrolment in the study. Ethical approval was given by the Faculty of Dentistry, University of Malaya Ethical Committee (reference number: DF RD1514/0047(P)) for the use of human individuals in this clinical study.
All teeth except the third molars were considered for the periodontal clinical measures by one calibrated examiners (ZN). Clinical parameters consisting of plaque score (PS) [22], bleeding on probing (BOP), pocket depth (PD) and clinical attachment loss (CAL) were measured at six sites for each tooth. The Intra Class Correlation Coefficient test was used to assess the intra examiner reliability which was 0.86 and 0.85 for PD and CAL respectively. Based on the clinical measurements and radiographs, the subjects were enrolled into the healthy, gingivitis or chronic periodontitis groups. In total, 30 subjects were diagnosed with moderate to severe chronic periodontitis according to the American Academy of Periodontology task force report on the update to the 1999 Classification of Periodontal diseases and conditions [23]. If the PD≥5mm and CAL ≥3-4, patients were considered as having moderate chronic periodontitis. While when the PD≥7mm and CAL >5mm, patients were considered as having severe chronic periodontitis.
Subjects in healthy and gingivitis groups displayed less than 3mm of CAL, PD not more than 4mm and no radiographic evidence of alveolar bone loss. Subjects with BOP less than 20% were considered as healthy (H) (n=28), and those with BOP more than 20% were considered as Gingivitis (G) (n=30). Each study group (H, G, and CP) was subdivided into three subgroups according to the participants’ ethnicity, in which the G and CP groups included Malays (n=10), Chinese (n=10) and Indians (n=10), while the H group included Malays (n=10), Chinese (n=9) and Indians (n=9).
Subgingival plaque collection
A total of 352 subgingival plaque samples were collected. Briefly, the upper posterior teeth excluding the third molar were selected initially for the subgingival plaque sample collections, which were taken from four sites of each healthy, gingivitis and chronic periodontitis subjects. If those teeth did not show PD≥5 mm and CAL≥3-4 mm in the chronic periodontitis patients, they were replaced by the adjacent teeth. After a gentle removal of supra gingival plaque, the test sites were air dried and kept dry using cotton rolls. Sterile paper points (size #30) were inserted into the selected area (periodontal pockets or gingival sulcus) for 30 seconds to obtain the subgingival plaque. Any paper point showing blood contamination was discarded. The paper point from each sampling site was immediately placed into a microfuge tube containing 1ml phosphate buffer saline. The samples for the qPCR analysis were then stored at -80 °C.
DNA extraction
The DNA extraction kit (DNeasy® Blood and Tissue Kit (50), Cat. No. 69504) was used for the bacterial DNA extraction. The manufacturing protocol was as follows: the samples were thawed for 20-30minutes and placed into a vortex for 30seconds and spun in a micro centrifuge for 10seconds. The paper points were then removed from the microfuge tubes and pelleted by refrigerated centrifuge at 13.2000rpm for 30 minutes at 4 °C and then the supernatant was discarded. After that 180μl of tissue lysis buffer (ATL) and 20μl of proteinase K were added to the pelleted cells followed by vortexing for 10 seconds and spinning for 10 seconds after which it was placed in a thermo mixer at 56 °C for 3hours for incubation to take place. After the samples were removed from the thermo mixer they were spun for 10 seconds. A 200μl of buffer (AL) was added to the tubes followed by vortexing for 10 seconds and spinning for 10 seconds and then incubated in the thermo mixers at 65 °C for 10 minutes’ samples then were spun for 10 seconds after being removed from the thermo mixer.
200μl of cold absolute ethanol was added to each of the samples and the mixture was transferred to the DNeasy mini spin column and centrifuged at 8000rpm for 1 minute and then the flow was discarded. A 500μl of washing buffer 1 (AW1) was added and centrifuged at 8000rpm for 1 minute and then the flow was discarded. A 500μl of washing buffer 2 (AW2) was then added and centrifuged at 8000rpm for 1 minute and the flow was then discarded. A new collection tube was placed and centrifuged at 13.2000rpm for 3 minutes for drying. The total bacterial DNA was then transferred to a new micro centrifugal tube and eluted with 35μl of elution buffer (AE), and then the DNA concentration and quality were measured using NANODROP 2000 spectrophotometer device and the total product was stored at -20 °C until it was time to do the real-time polymerase chain reaction analysis.
Preparation of P.gingivalis, T. forsythia, T. denticola and A. actinomycetemcomitans standard curve
The manufacturer protocol (PrimerDesignTM Ltd Genesig) was followed to obtain the standard curve for the four types of bacteria (P.gingivalis, T. forsythia, T. denticola and A. actinomycetemcomitans). The procedure involved a six series dilution from 2x105 to 2x100 in which the lyophilised positive control was added with 500μl of RNAse/DNAes free water. Then the following steps were followed: A 900μl of RNAse/DNAes free water was pipetted into five tubes (1.5ml Eppendorf tubes) and labelled 2 to 6. Then a 100μl of positive control template which contained a high copy number template was pipetted into tube No. 2 and vortexed thoroughly. After that, a 100μl was pipetted from tube 2 into tube 3 and also vortexed thoroughly. These steps were repeated for tubes 4 to 6 in order to complete the series of dilution.
Bacterial quantification by real-time PCR
Primer DesignTM Ltd Genesig kits were used for the quantification of P.gingivalis, T. forsythia, T. denticola and A. actinomycetemcomitans in which the head shock protein 60 genes were used to detect and quantify the P.gingivalis, T. forsythia, T. denticola and A. actinomycetemcomitans genome. The Fast-Real- Time PCR System (Applied Biosystems, Foster City, CA, USA) was used for the quantification of the bacterial genomic DNA according to the manufacturer protocol. The reaction mix which includes sufficient reactions for the negative control and standard curve wells was prepared. Then a 15μl of this mix was pipetted into each well according to the real-time PCR plate set up. PCR was performed in a total volume of 20μl consisting of 5μl of genomic DNA, Ten μl of lyophilised Master mix (Primer Design Ltd OasigTM 2X), 4μl of RNAse/DNAes free water and 1μl of each bacterial species primer/probe mix. Five μl of RNAse/DNAes free water was added to the negative control well making the final volume become 20μl in each well. Finally, the quantitative PCR was done on the Fast Real-Time PCR using the manufacturer PCR conditions as follows: initial denaturation for enzyme activation for 15 minutes at 95 °C, followed by 50 cycles for denaturation (10 seconds at 95 °C) and data collection (60 seconds at 60 °C). The quantity of P.gingivalis, T. forsythia, T. denticola and A. actinomycetemcomitans DNA was generated by the Fast Real-Time PCR System.
GCF collection
GCF samples were collected from 528 sites. The collection of the GCF samples was according to the method of Tezel et al, [24]. The GCF samples collected from the same teeth were also chosen to collect the subgingival plaque samples. The interproximal crevicular sites in the upper posterior region excluding the third molar were selected and isolated with cotton rolls and the tooth surfaces were air dried to avoid contamination with saliva. The GCF samples were collected using absorbent paper strip (Periopaper®, ProFlow Inc., Amityville, NY, USA) which was placed into the sulcus/pocket until mild resistance was felt and the strip was held in place for 30 seconds. Strips contaminated by saliva or blood were excluded and another site was chosen to obtain the sample. A total of six absorbent paper strips were obtained from each subject. All the strips with GCF were immediately placed into a sterile polypropylene tube and kept at -80 °C. Then a 200μl of phosphate buffer saline was added to each tube which was shaken gently for 1 hour at room temperature. The strips were then removed and the fluid was stored at -80 °C until assayed by ELISA.
ELISA
The ELISA kit Hycult® Biotech Human LL-37 (Cat. No. HK321- 01) was used for the quantitative determination of human gingival crevicular fluids LL-37. The ELISA kit protocol was followed to calculate the mean absorbance of samples. All reagents were placed at room temperature (20-25 °C) before analysis. A 100μl of standards and samples were transferred into appropriate wells and the tray was covered and tapped to eliminate any air bubbles. The plate was then incubated for 1 hour at room temperature. After incubation, the plates were washed four times with a 200μl wash/ dilution buffer each time and then a 100μl of diluted tracer was added to each well using the same pipetting order. The plate was then covered, tapped and incubated for 1 hour. After incubation, the plate was again washed four times, as previously done, after which 100μl of diluted streptavidin-peroxidase was added to each well and the tray was again covered and incubated for 1 hour. The washing procedure was repeated for four times, after which a 100μl of tetramethylbenzidine (TMB) substrate was added to each well using the same pipetting order and the tray was incubated for a ½hour at room temperature. Finally, a 100μl of the stop solution was added to each well with the same sequence and then the plate was read within 30 minutes after the addition of stop solution at 450nm using a Tecan Genios microtitre plate reader (Tecan, Reading, UK) using the Magellan software. A standard curve was drawn in order to calculate the LL-37 concentration.
Statistical analysis
Statistical analyses were performed using the parametric and non-parametric methods. The normality of data was checked by Shapiro-wilk test. The GCF human cathelicidine LL-37 was analysed parametrically with the one-way ANOVA test and the differences among the three groups were analysed with a post hoc Bonferroni test. The clinical periodontal parameters (PS, BOP, PD, and CAL) and subgingival plaque pathogens were analysed nonparametrically with the Kruskal-Wallis test and the differences among the three groups were analysed with a post hoc Dunn test. The two-way ANOVA test was used to analyse the ethnic subgroup data. The correlation between the variables was achieved by using the Spearman rho rank test. All the data were analysed using the IBM SPSS version 20 software package.
Result
Clinical and demographic findings
The clinical and demographic data of the subjects who contributed to this study are shown in Table 1. The average mean age was significantly higher in the CP group compared to the H and G groups (p<0.05). The mean PS and BOP scores were significantly higher in the G and CP groups compared to the H group (p<0.05), in which there was no statistically significant difference between the G and CP groups (p>0.05). The CP group had significantly higher mean PD and CAL scores compared with the G and H groups (P <0.05). The H and G groups had no significant difference in the mean PD score (p>0.05) but similar CAL scores (Table 1).
Table 1:Clinical periodontal parameters and demographic characteristics of the study population.
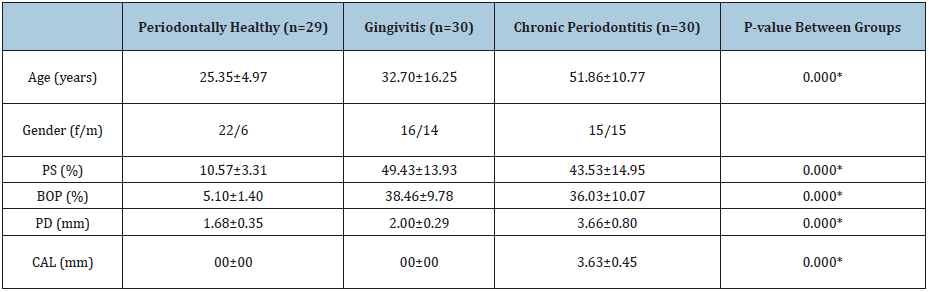
*P<0.05; significant difference.
Laboratory findings
Subgingival pathogens: The total counts of P.gingivalis and T.denticola are presented in Figures 1A and 1C, respectively. There was a high statistically significant difference (p<0.05) in the values between the study groups. This difference was mainly between the H and CP groups (p<0.05), and G and CP groups (p<0.05). However, the difference between the H and G groups was not significant (p>0.05). The counts of T.forsythia showed a high significant difference (p<0.05) between the H, G and CP groups (Figure 1B). This difference was mainly between the H and CP groups, and G and CP groups (p<0.05), and there was a slight significant difference between the H and G groups (p<0.05). There was a statistically significant difference in the count of A.a between the study groups (P<0.05) as shown in Figure 1D. The multi comparisons between the groups using the post hoc Dunn test showed that the statistical difference was between the G and CP groups only (p <0.05). There were no statistical differences in the quantity of P.gingivalis, T.forsythia, T.denticola and A.a between the three ethnic subgroups of the H, G and CP groups (P>0.05) (Table 2) (Figure 1).
A. P.gingivalis counts in the study groups.
B. T.forsythia counts in the study groups.
C. T.denticla counts in the study groups.
D. A.a counts in the study groups. (*) represents extreme values and the circle represent the outliers.
Correlations of subgingival pathogens with the clinical periodontal parameters: The P. gingivalis counts in the subgingival plaque were not correlated to PS and BOP (P>0.05), but there was a positive correlation between the P. gingivalis counts with PD and CAL (rs=0.442, rs=0.494, respectively). The T. forsythia counts were found to be positively correlated to all the clinical periodontal parameters (PS, BOP, PD and CAL) in which rs values were as follows: 0.347, 0.415, 0.619 and 0.660, respectively. The T. denticola counts were not correlated with PS and BOP (P>0.05) but it was positively correlated with PD and CAL (rs=0.443 and rs=0.5, respectively). The A.a counts were not correlated to PS and BOP (P>0.05) but positively correlated to PD and CAL (rs=0.345, rs=0.327) (Table 3).
Figure 1:Periodontal pathogens counts in the H, G, and CP groups
A) P. gingivalis counts in the study groups. B) T. forsythia counts in the study groups. C) T. denticla counts in the study groups. D) A. a counts in the study groups. (*) represents extreme values and the circle represent the outliers.
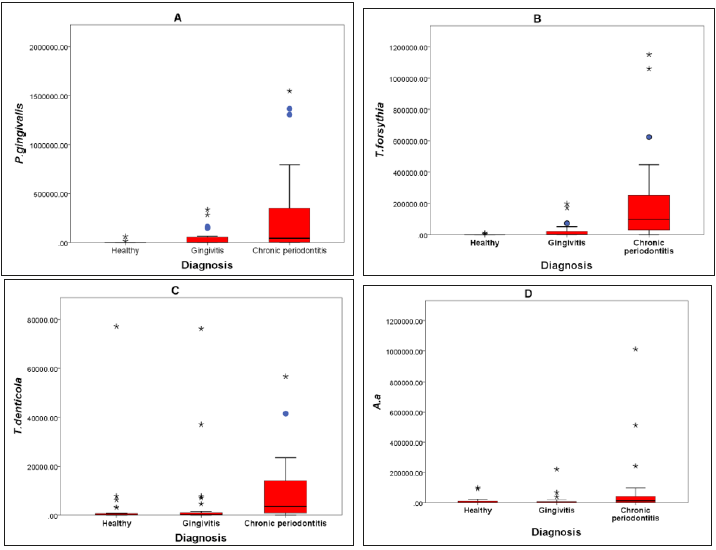
Table 2:Red complex species and A.a mean and standard deviation in the different ethnic groups (10×4).
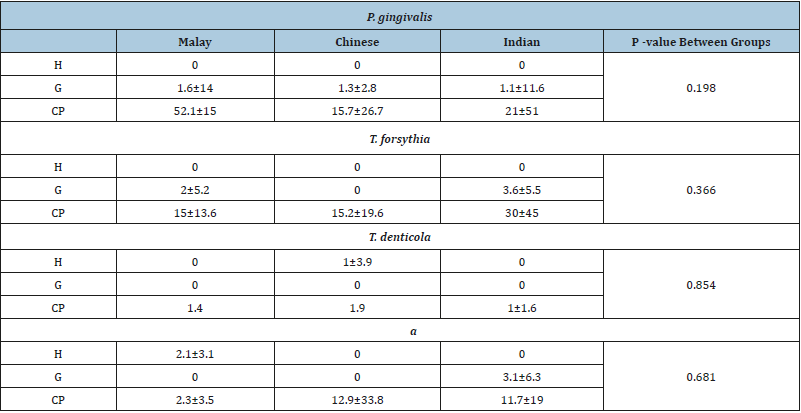
P<0.05; significant difference.
Table 3:Correlations of subgingival pathogens with the clinical periodontal parameters.
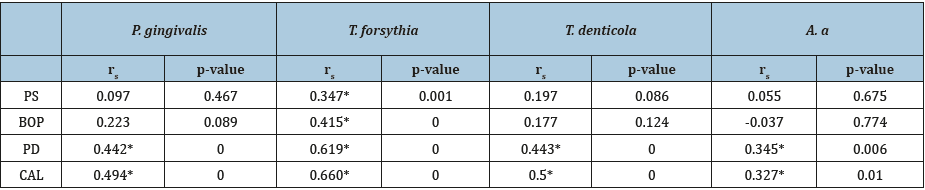
*Correlation significance at the 0.01 level (two-tailed).
Human cathelicidine LL-37 in GCF: Figure 2 shows the distribution of human cathelicidine LL-37 levels in the study groups; the LL-37 levels were significantly higher in the CP group compared to the H and G groups (p<0.05). There was no statistical difference between the H and G groups (p>0.05). The detection rate of LL- 37 in GCF was 100% in CP group, while in the H and G groups the detection rate was 78.5% and 93.3%, respectively. The differences in the LL-37 levels between the three ethnic subgroups in the H, G, and CP groups were determined and there was a significant difference (p<0.05) as shown in Table 4, the post hoc Bonferroni test showed that there was a minor significant difference (p= 0.041) between the Malay and Chinese ethnic groups only, while there were no statistical differences (p>0.05) between the Malay and Indian or Chinese and Indian ethnic groups.
Figure 2:LL-37 concentrations (ng/mL) between different races.
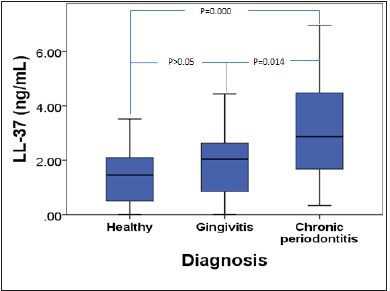
Table 4:LL-37 concentrations (ng/mL) between different races.
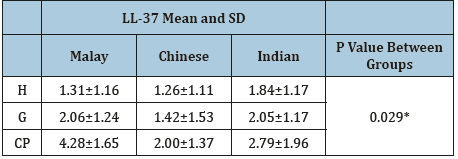
*P<0.05; significant difference.
Correlations of the LL-37 levels with the clinical periodontal parameters and periodontal pathogens:There were no correlations between the LL-37 levels in the GCF with the PS and BOP but the LL-37 levels were positively correlated with PD and CAL. The LL-37 levels were positively correlated with the P. gingivalis, T. forsythia, T. denticola and A. actinomycetemcomitans quantity (rs=0.337, rs=0.365, rs=0.371 and rs=0.296) respectively (Table 5).
Table 5:Correlations of the LL-37 levels with the clinical periodontal parameters and periodontal pathogens.
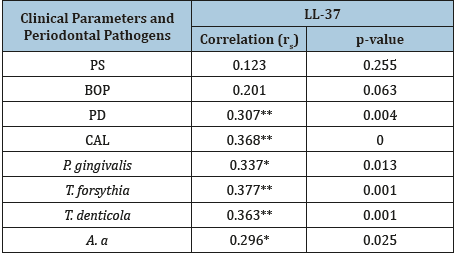
**Correlation significant at the 0.01 level (two-tailed).
*Correlation significant at the 0.05 level (two-tailed).
Discussion
This is a cross sectional comparative study of the cathelicidine LL-37 levels in the gingival sulcus /periodontal pockets taken from the GCF samples among the different ethnic groups in the Malaysian population. To the best of our knowledge, this is the first study that compared the GCF LL-37 levels in three ethnic groups within the same population. The aim of this study was to determine and correlate the levels of human cathelicidine (LL-37) and subgingival periodontal red complex microorganisms and A.a pathogens in the periodontal health and disease among three ethnic groups in Malaysia. Many studies have demonstrated that the composition of subgingival pathogens amongst different populations in the world is important to provide information into the bacterial changes during the different stages of periodontal disease progression that occurs in individuals from different geographical regions [25]. This study observed that the red complex species strongly colonized the subgingival microbiota in the chronic periodontitis patients when compared to healthy subjects or patients with gingivitis.
This finding is consistent with other studies in which periodontopathic bacteria were less detected in healthy subjects [26-28] and patients with gingivitis [29,30]. The pathological deepening of the gingival sulcus and increased amount of dental plaque biofilm harbour further counts of periodontopathic bacteria [31]. Our findings also showed that the counts of A.a in the Malaysian population were not correlated to the periodontal disease status, in which there was no significant difference in the counts between the H and CP groups and slight difference between the G and CP groups. This finding is in accordance with a study done on the Thai population that also showed no correlation between the A.a and periodontal disease status by reporting that the prevalence of A.a in the moderate to severe CP patients (35%) was less than the prevalence of A.a in the mild CP group (45%) [28]. Furthermore, a study done on Japanese population was unable to confirm the significant difference between the various types of periodontal diseases. The study showed that there was a low prevalence of A.a in the localized aggressive periodontitis, generalized aggressive periodontitis, and CP in the study groups [32]. Data discrepancy regarding the quantification and prevalence of the subgingival periodontal pathogens in patients with different stages of periodontal disease among different populations maybe due to numerous reasons which among them are the differences in the followings: classification of periodontal disease, data analysis methods, techniques used for sample analysis [27], demographic features of the study population [33], ethnicity, cultural factors [31], socioeconomic status and geographic locations [28].
Only a few studies have been able to investigate the differences between ethnic groups within the same geographical location [34,35] or more than one population under the same investigational conditions within similar research abilities [36]. One of the main goals of the present study was to explore if the genetic diversity among different ethnic groups has an effect on subgingival pathogens distributed in different stages of the periodontal disease. The results of our study showed that there was no marked effect of the genetic diversity on the subgingival pathogens counts. There were no statistical differences in the T. forsythia, T. denticola, P. gingivalis and A.a counts between Malay, Chinese and Indian populations in the H, G, and CP groups. This result is in accordance with a study performed by Sirinian et al, [34] who investigated the presence of T. forsythia, T. denticola, P. gingivalis and A.a in the saliva samples taken from young subjects in the Caucasians, Hispanics and Asian-American ethnic groups resided in the Los Angeles, where they found that the periodontopathic bacteria were not correlated to the ethnicity but related to the education level of the mother, gender and environmental factor [34]. In a different study conducted by Haffajee et al, [36] to explore the composition of the subgingival plaque bacteria in the CP patients from four countries (USA=115 subjects; Sweden=101 subjects; Chile=26 subjects; Brazil=58 subjects) using almost standardized methods in the sample collections and analysis, a marked difference in the mean proportion of many types of subgingival plaque bacteria was reported. This mainly contributed to the differences in probing depths of the sampled sites between the groups, some groups receiving periodontal treatment or systemic antibiotics, in addition to diet, culture, genetic background and differences in socio-economic status 36. In humans, LL-37 appears to play a major role in protecting the periodontal tissue against dental plaque microbes [37], although the concentration of LL-37 is insufficient to prevent the initiation and progression of periodontal diseases. This especially presents in genetic disorders like the Morbus Kostmann syndrome, which is a severe congenital disease that causes a deficiency of LL-37 [38] and Papillon-Lefe`vre syndrome where there is also a lack of LL-37 [39] which plays a key role in the development of severe periodontal destruction in these patients.
The stimulation of periodontal pathogens during inflammation leads to the expression of LL-37 from the gingival epithelial cells [40], salivary glands [41], monocytes [42] and mainly from neutrophils [37]. In the present study, the cathelicidine LL-37 levels from the GCF samples significantly elevated in the CP patients compared to the H subjects. This finding is consistent with other studies conducted by [37,43,44]. The significant differences in the LL-37 levels were also found between the G patients and other study groups whereby the same findings were demonstrated by Türkoglu et al, [44] in which the CP patients were reported to have more LL-37 levels than the G patients who had elevated LL- 37 levels compared to the H subjects. Additionally, in this study, the LL-37 levels were found to be positively correlated to the probing depths and clinical attachment loss which are considered the most accurate clinical parameters together with radiographic interpretation in determining the severity of periodontal diseases. These findings are consistent with a study by Hosokawa et al, [40] who measured the LL-37 levels in the gingival tissue from different gingival sulcus/periodontal pocket depths and reported a positive correlation between the LL-37 and pocket depths [40]. Türkoglu et al, [44] also stated such positive correlation between the LL- 37 and pocket depths and they contributed this relationship to the stimulation of inflammatory response during the periodontal disease progression by dental plaque.
This stimulation leads to neutrophils migration in the periodontal pocket which causes LL-37 to be released from the stimulated neutrophils [44]. These findings of cathelicidine LL-37 in the GCF expression relative to the periodontal disease severity may assist in the prediction and diagnosis of different periodontal diseases as LL-37 plays an important role in the innate immune response and thus using LL-37 as a useful biomarker for the progression of periodontal diseases. This is especially because cathelicidine LL-37 in the GCF was also detected in healthy subjects (78.5%) and the G and CP groups (93.3%, 100%, respectively) in different concentrations. This could be due to the neutrophils stimulation by the many periodontal pathogens leading to LL- 37 secretions through the junctional epithelium in the gingival sulcus. The neutrophil sensitivity to a minor bacterial stimulus as in healthy subjects could be useful in the early prediction and monitoring of periodontal disease through the monitoring of LL-37 levels in GCF. The occurrence of LL-37 related with neutrophil only in the periodontally healthy subjects suggested that neutrophils are the essential factor of the LL-37 release in the healthy periodontium [37]. Furthermore, the present study demonstrated a positive correlation between the LL-37 levels and quantity of the T. forsythia, T. denticola, P. gingivalis and A.a in subgingival plaque. These microbes are considered to be the most pathogenic for the initiation and progression of periodontal diseases. The same correlation was discovered by [37] who demonstrated that the unprocessed cathelicidine and mature LL-37 levels in the GCF were positively correlated to the T. forsythia, T. denticola and P. gingivalis pathogens. This positive correlation between the LL-37 levels and red complex species and A.a explained the elevation of LL-37 levels in accordance with the severity of periodontal disease which supported our study of using LL-37 as a biomarker for periodontal diseases.
The variations in the expressions of GCF LL-37 levels in three ethnic groups within the Malaysian population were investigated in this study in an attempt to explore the effects of genetic discrepancies on the cathelicidine LL-37 levels. The statistical difference (p=0.41) was detected between the Malay and Chinese groups and there was no statistical difference between other study groups. This slight difference between the Malay and Chinese groups may be due to the small sample size (n=10) of the ethnic subgroups enrolled in each of the study groups (H, G, and CP). Since the study samples were collected from the same geographical region and had similar nutritional and environmental conditions, the authors were in the opinion that demographic and socioeconomic variations between the study groups may have played a role in this slight difference between the Malay and Chinese groups.
Conclusion
This study showed that the red complex species counts within the same study population can be useful in the differentiation between various stages of periodontal disease progression. The genetic backgrounds of the subjects had no noticeable effect on the periodontal pathogen expression during the periodontal disease progression. The results of this study highlighted the role of human cathelicidine (LL-37) in the innate immunity against periodontal inflammation. This study showed that there was a positive correlation between the LL-37 levels and periodontal pathogens which play a major role in the initiation and progression of chronic periodontitis. LL-37 in GCF may be considered as a relevant biomarker for the periodontitis diseases. The GCF LL- 37 levels may support the clinical periodontal parameters in the diagnosis of periodontal diseases. However, the limitations of this study were the small sample size of the different ethnic groups and difficulties in matching the demographic criteria of the study groups, particularly the age variations between the CP groups and other study groups. Further studies are needed to measure the LL- 37 levels in different ethnic groups with a larger sample size so that the genetic differences between subjects and its impact on the LL-37 concentrations can be explored more thoroughly taking into consideration the demographic data matching as much as possible.
Acknowledgement
This work was supported by the University of Malaya’s High Impact Research with Malaysian Ministry of Higher Education (HIRMOHE) research grant UM.C/625/1/HIR/MOHE/SC/08 (account number F000008-21001) under the Principal Investigator Koshy Philip.
Compliance with Ethical Standards
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Conflict of Interest
The authors declare that they have no conflict of interest.
Informed Consent
A. Informed consent was obtained from all individual participants included in the study.
References
- Armitage GC, Cullinan MP (2010) Comparison of the clinical features of chronic and aggressive periodontitis. Periodontology 2000 53(1): 12-27.
- Teles RP, Haffajee AD, Socransky SS (2006) Microbiological goals of periodontal therapy. Periodontology 2000 42(1): 180-218.
- Heller D, Varela VM, Torres MCB, Feres‐Filho EJ, Colombo APV (2011) Impact of systemic antimicrobials combined with anti‐infective mechanical debridement on the microbiota of generalized aggressive periodontitis: a 6‐month RCT. J Clin Periodontol 38(4): 355-364.
- Rylev M, Kilian M (2008) Prevalence and distribution of principal periodontal pathogens worldwide. J Clinical Periodontology 35(8 Suppl): 346-361.
- Socransky SS, Haffajee AD (2002) Dental biofilms: difficult therapeutic targets. Periodontology 2000 28(1): 12-55.
- Colombo APV, Teles RP, Torres MC, Souto R, Rosalém WJ, et al. (2002) Subgingival microbiota of Brazilian subjects with untreated chronic periodontitis. J Periodontol 73(4): 360-369.
- Shaddox LM, Walker C (2009) Microbial testing in periodontics: value, limitations and future directions. Periodontology 2000 50(1): 25-38.
- Socransky S, Haffajee A, Cugini M, Smith C, Kent RL (1998) Microbial complexes in subgingival plaque. J Clinical Periodontol 25(2): 134-144.
- Holt SC, Ebersole JL (2005) Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: the ‘red complex’, a prototype polybacterial pathogenic consortium in periodontitis. Periodontology 2000 38(1): 72-122.
- Thurnheer T, Belibasakis GN, Bostanci N (2014) Colonisation of gingival epithelia by subgingival biofilms in vitro: role of “red complex” bacteria. Arch Oral Biol 59(9): 977-986.
- Li Y, Shibata Y, Zhang L, Kuboyama N, Abiko Y (2011) Periodontal pathogen Aggregatibacter actinomycetemcomitans LPS induces mitochondria-dependent-apoptosis in human placental trophoblasts. Placenta 32(1): 11-19.
- Kang J, de Brito Bezerra B, Pacios S, Andriankaja O, Li Y, et al. (2012) Aggregatibacter actinomycetemcomitans infection enhances apoptosis in vivo through a caspase-3-dependent mechanism in experimental periodontitis. Infect Immu 80(6): 2247-2256.
- Wang G, Mishra B, Epand RF, Epand RM (2014) High-quality 3D structures shine light on antibacterial, anti-biofilm and antiviral activities of human cathelicidin LL-37 and its fragments. Biochim Biophys Acta (BBA)-Biomembranes 1838(9): 2160-2172.
- Gorr SU, Abdolhosseini M (2011) Antimicrobial peptides and periodontal disease. J Clin Periodontol 38(Suppl 11): 126-141.
- Tonetti MS, Chapple IL (2011) Biological approaches to the development of novel periodontal therapies-consensus of the Seventh European Workshop on Periodontology. J Clinical Periodontol38(Suppl 11): 114-118.
- Vandamme D, Landuyt B, Luyten W, Schoofs L (2012) A comprehensive summary of LL-37, the factotum human cathelicidin peptide. Cellular Immunol 280(1): 22-35.
- Cowland JB, Johnsen AH, Borregaard N (1995) hCAP‐18, a cathelin/pro‐bactenecin‐like protein of human neutrophil specific granules. FEBS Lett 368(1): 173-176.
- Sørensen OE, Follin P, Johnsen AH, Calafat J, Tjabringa GS, et al. (2001) Human cathelicidin, hCAP-18, is processed to the antimicrobial peptide LL-37 by extracellular cleavage with proteinase 3. Blood 97(12): 3951-3959.
- Yang D, Oppenheim J (2009) Alarmins and antimicrobial immunity. Medical Mycology 47(sup1): S146-S153.
- Ertugrul A, Sahin H, Dikilitas A, Alpaslan N, Bozoğlan A, et al. (2014) Gingival crevicular fluid levels of human beta‐defensin‐2 and cathelicidin in smoker and non‐smoker patients: a cross‐sectional study. J Periodontal Res 49(3): 282-289.
- McCrudden MT, Orr DF, Yu Y, Coulter WA, Manning G, et al. (2013) LL‐37 in periodontal health and disease and its susceptibility to degradation by proteinases present in gingival crevicular fluid. Journal of clinical periodontology 40(10): 933-941.
- O Leary TJ, Drake RB, Naylor JE (1972) The plaque control record. Journal of periodontology 43(1): 38.
- Periodontitis (2015) American academy of periodontology task force report on the update to the 1999 classification of periodontal diseases and conditions. J Periodontol 86(7): 835-838.
- Tezel A, Yucel O, Orbak R, Kara C, Kavrut F, et al. (2005) The gingival crevicular fluid ciprofloxacin level in subjects with gingivitis and periodontitis, and its effects on clinical parameters. J Periodontal Research 40(5): 395-400.
- Ximenez Fyvie LA, Almaguer Flores A, Jacobo Soto V, Lara Cordoba M, Sanchez Vargas LO, et al. (2006) Description of the subgingival microbiota of periodontally untreated Mexican subjects: chronic periodontitis and periodontal health. J Periodontol 77(3): 460-471.
- Tomita S, Komiya-Ito A, Imamura K, Kita D, Ota K, et al. (2013) Prevalence of Aggregatibacter actinomycetemcomitans, Porphyromonas gingivalis and Tannerella forsythia in Japanese patients with generalized chronic and aggressive periodontitis. Microb Pathog 61-62: 11-15.
- Dogan B, Antinheimo J, Çetiner D, et al. (2003) Subgingival microflora in Turkish patients with periodontitis. J Periodontol 74(6): 803-814.
- Wara aswapati N, Pitiphat W, Chanchaimongkon L, Taweechaisupapong S, Boch J, et al. (2009) Red bacterial complex is associated with the severity of chronic periodontitis in a Thai population. Oral Dis 15(5): 354-359.
- Tanner A, Maiden M, Macuch P, Murray L, Kent R (1998) Microbiota of health, gingivitis, and initial periodontitis. J Clin Periodontol 25(2): 85-98.
- Ashimoto A, Chen C, Bakker I, Slots J (1996) Polymerase chain reaction detection of 8 putative periodontal pathogens in subgingival plaque of gingivitis and advanced periodontitis lesions. Oral Microbiol Immunol 11(4): 266-273.
- Botero JE, Contreras A, Lafaurie G, Jaramillo A, Betancourt M, et al. (2007) Occurrence of periodontopathic and superinfecting bacteria in chronic and aggressive periodontitis subjects in a Colombian population. J Periodontol 78(4): 696-704.
- Takeuchi Y, Umeda M, Ishizuka M, Huang Y, Ishikawa I (2003) Prevalence of periodontopathic bacteria in aggressive periodontitis patients in a Japanese population. J Periodontol 74(10): 1460-1469.
- Braga RR, Carvalho MAR, Bruña Romero O, Teixeira RE, Costa JE, et al. (2010) Quantification of five putative periodontal pathogens in female patients with and without chronic periodontitis by real-time polymerase chain reaction. Anaerobe 16(3): 234-239.
- Sirinian G, Shimizu T, Sugar C, Slots J, Chen C (2002) Periodontopathic bacteria in young healthy subjects of different ethnic backgrounds in Los Angeles. J Periodontol 73(3): 283-288.
- Ellwood R, Worthington H, Cullinan M, Hamlet S, Clerehugh V, et al. (1997) Prevalence of suspected periodontal pathogens identified using ELISA in adolescents of differing ethnic origins. J Clinical Periodontol 24(3): 141-145.
- Haffajee A, Bogren A, Hasturk H, Feres M, Lopez N, et al. (2004) Subgingival microbiota of chronic periodontitis subjects from different geographic locations. J Clin Periodontol 31(11): 996-1002.
- Puklo M, Guentsch A, Hiemstra P, Eick S, Potempa J (2008) Analysis of neutrophil‐derived antimicrobial peptides in gingival crevicular fluid suggests importance of cathelicidin LL‐37 in the innate immune response against periodontogenic bacteria. Oral Microbiol Immunol 23(4): 328-335.
- Pütsep K, Carlsson G, Boman HG, Andersson M (2002) Deficiency of antibacterial peptides in patients with morbus Kostmann: an observation study. The Lancet 360(9340): 1144-1149.
- de Haar SF, Hiemstra PS, van Steenbergen MT, Everts V, Beertsen W (2006) Role of polymorphonuclear leukocyte-derived serine proteinases in defense against Actinobacillus actinomycetemcomitans. Infect Immun 74(9): 5284-5291.
- Hosokawa I, Hosokawa Y, Komatsuzawa H, Goncalves RB, Karimbux N, et al. (2006) Innate immune peptide LL‐37 displays distinct expression pattern from beta‐defensins in inflamed gingival tissue. Clin Exp Immunol 146(2): 218-225.
- Tao R, Jurevic RJ, Coulton KK, Tsutsui MT, Roberts MC, et al. (2005) Salivary antimicrobial peptide expression and dental caries experience in children. Antimicrob Agents Chemother 49(9): 3883-3888.
- Scott MG, Davidson DJ, Gold MR, Bowdish D, Hancock RE (2002) The human antimicrobial peptide LL-37 is a multifunctional modulator of innate immune responses. J Immunol 169(7): 3883-3891.
- Makeudom A, Kulpawaropas S, Montreekachon P, Khongkhunthian S, Sastraruji T, et al. (2014) Positive correlations between hCAP18/LL‐37 and chondroitin sulphate levels in chronic periodontitis. J Clin Periodontol 41(3): 252-261.
- Türkoglu O, Emingil G, Kütükçüler N, Atilla G (2009) Gingival crevicular fluid levels of cathelicidin LL-37 and interleukin-18 in patients with chronic periodontitis. J Periodontol 80(6): 969-976.
© 2019 Koshy Philip. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)













.bmp)
.jpg)
.png)
.jpg)














.png)

.png)



.png)






