- Submissions

Full Text
COJ Reviews & Research
Differentiation between Snake and Bee Venom using Fluorescence Spectroscopy and Computational Approach
Alla M Hashkel1, Enas A Sadawe1, Asia Mohamed1, Nesren M Magel1, Ibrahim A Mrema1, Salah M Bensaber1, Fathi M Sherif2, Massaud Salem Maamar3, Amira Abdulhakim Elmaghrbi3, Anton Hermann4 and Abdul M Gbaj1*
1Department of Medicinal Chemistry, University of Tripoli, Libya
2Department of Pharmacology, University of Tripoli, Libya
3Department Zoology, Tripoli University, Libya
4Department of Biosciences, University of Salzburg, Austria
*Corresponding author: Abdul M Gbaj, Department of Genetics and Biochemistry, University of Tripoli, Libya
Submission: May 21, 2018; Published: July 10, 2018

ISSN 2639-0590Volume1 Issue2
Abstract
Phospholipase A2 (PLA2) represents a major venom component of snakes and bees and exhibits a broad range of biological effects including myotoxicity, neurotoxicity, hemolysis, cardiotoxicity, anticoagulant and antiplatelet activities. Melittin, is the main component and the major pain producing substance of honeybee venom. The aim of the present study was to differentiate between snake and bee venoms using Aqueous Olive Leaf Extract (AOLE) employing fluorescence techniques. Tryptophan, which is contained in both snake and bee venoms is fluorescent at UV wavelength and hence widely used as a tool to monitor conformational changes in proteins and to draw inferences regarding local structure and dynamics. Fluorescence spectroscopy and molecular modeling have been used to analyze enzyme activity in the absence and presence of AOLE and to verify potential binding of AOLE components to the enzyme. Changes in the fluorescence intensities with blue and red shifts were obtained with bee and snake venoms, respectively. Binding of AOLE constituents near the active site of the enzyme could be evidenced and possible modes of interaction are discussed. The fluorescence method used was rapid and sensitive and was able to differentiate between snake and bee venoms utilizing AOLE.
keywordsSnakes and Bees; Toxicity; Venom; Fluorescence Spectroscopy; AOLE constituents
Abbreviations: PLA2: Phospholipase A2; AOLE: Aqueous Olive Leaf Extract; ROS:Reactive Oxygen Species; RMS: Root Mean Square
Introduction
Libya and surrounding countries are inhabited by numerous venomous snakes of medical importance. Among them are the families Viperidae, Hydrophiidae, Elapidae and Atractaspididae. Cerastes cerastes mainly inhabits North Africa (Libya, Sudan, Algeria, Tunisia etc.), and is sympatric with another Asian species Cerastes gasperettii found in (Jordan, Iran, Palestine, and Iraq). Venoms from snakes within the family Viperidae are complex biochemical mixtures that function to immobilize the prey and initiate digestion. Envenomation by viperid and crotalid snakes is typically characterized by hemorrhage and necrosis that can cause lasting tissue damage [1] if the individual survives the predation. Cerastes cerastes venom contains many enzymes showing proteolytic activity and causes multiple kinds of intoxications [2]. The toxicities cause substantial physiopathological changes in the liver, skin and heart. Phospholipases A2 (PLA2) from Cerastes cerastes for example, has been associated with numerous toxicities including neurotoxicity, nephrotoxicity, lung toxicity, hepatotoxicity, and cardiotoxicity [3-5]. The lethal effect of snake venom mainly results from its active ingredients such as phospholipase A2 (PLA2). Phospholipid hydrolysis by PLA2 releases arachidonic acid whose metabolism results in the formation of potentially toxic Reactive Oxygen Species (ROS) and lipid peroxides [5,6]. The increase in the activity of liver enzymes indicating the damage of heart, liver and other organs could be attributed to the synergistic action of the venom components [2,5].
Hemorrhagic effects of snake venoms are commonly measured by irritation of the skin, a test developed by Kondo et al. using laboratory animals such as rabbits, rats, or mice [7,8]. In other cases measurements of the hemoglobin content in muscle or skin tissue exposed to the venom were used [1]. Unfortunately, these methods require large numbers of animals, extensive training and expertise in the methodology and yields highly variable results. Fluorescent substrates have already been used successfully to quantify venom endopeptidase activities and might provide a technique to larger scale analysis required for comparative studies of venoms and prey resistance [9]. The aim of this work is to validate a rapid and sensitive fluorescence method that can differentiate between snake and bee venoms using AOLE. Cerastes cerastes (Viperidae) venom is barely toxic, although it is reported to be similar in venom toxicity of Echis. Envenomation typically causes haemorrhage, necrosis, swelling, haematuria, nausea, and vomiting. C. cerastes contains a high content of phospholipase A2 which causes myotoxicity and cardiotoxicity [10]. Studies of venom from both C. cerastes and C. Vipera showed a total of eight venom fractions, the most powerful of which has haemorrhagic activity. Venom yields vary, with ranges of 19-27mg to 100mg of dried venom being reported for venom toxicity [10,11]. The LD50 values of 0.4mg/kg were applied Intravenous (IV) or 3mg/kg Subcutaneous (SC). The lethal dose for humans is 40-50mg [12].
Honey bee venom is a mixture of enzymes and peptides, a colorless and bitter tasting liquid. Enzymes in the bee venom comprise phospholipase A2, phospholipase B, α-glucosidase, hyaluronidase and phosphatase. In addition, bee venom contains peptides such as apamin, melittin, mast cell degranulating peptide, adolapin, tertiapin, melittin F, secapin and cardiopep [13-16]. Tryptophan (Trp) which is present in phospholipase A2 and melittin is a naturally occurring amino acid, which exhibits fluorescence emission properties that are dependent on the polarity of the local environment around the Trp side chain [17].
Materials and Methods
All experiments were conducted in Tris buffer (0.01M Tris, 0.1M NaCl, at pH 7.4). Glass-distilled deionized water and analytical grade reagents were used throughout experiments. pH values of solutions were measured with a calibrated Jenway pH-meter model 3510 (Staffordshire, UK). All buffer solutions were filtered through Millipore filters (Millipore, UK) of 0.45mm pore diameter
a. Absorbance spectra: Absorbance spectra were measured on a Jenway UV-visible spectrophotometer, model 6505 (London, UK) using quartz cells of 1.00cm path length. The UV-Vis absorbance spectra were recorded in the 200-500nm range, and spectral bandwidth of 3nm. For the final spectrum of each solution analyzed baseline subtraction of the buffer solution was performed. The protein content of venoms samples was determined by the spectrophotometric method of Markwell et al. [18]. Bovine serum albumin (BSA, Sigma) was used for standard assays.
b. Fluorescence spectra: Fluorescence emission and excitation spectra were measured using a Jasco FP-6200 spectrofluorometer (Tokyo, Japan) using fluorescence 4-sided quartz cuvettes of 1cm path length. The automatic shutter-on function was used to minimize photo bleaching of the sample. The selected wavelength chosen provided aggregate excitation of tryptophan and tyrosine residues. The emission spectrum was corrected for background fluorescence of the buffer.
Preparation of aqueous olea europaea leave extracts
Leaves of olive trees (Olea europaea) were collected from the Novellien zone, Tripoli Centre, Tripoli, Libya during April 2018. The leaves (5gm) were cleaned and washed with distilled water and dried at a room temperature of 25 °C for about 20 minutes. Dried leaves were grinded in a homogenizer (HO4A Edmund Buhler GmbH, UK) along with 15ml of distilled water. The resulting aqueous solution was filtered using a Millipore filter (0.45μm, GHD Acrodisc GF, UK).
Venoms
Snake (Cerastes cerastes, Macrovipera deserti, Echis pyramidum) and bee (A. m. scutellata) venoms were extracted by manual stimulation and were obtained in liquid and semisolid forms, respectively, from the Zoology Department, Faculty of Science, Tripoli University (Libya) and stored at -20 °C until use. Venoms were added to 2ml of 0.01 M Tris, 0.1 M NaCl at pH 7.4.
Molecular docking
The computational studies were performed in order to elucidate the binding and the interactions responsible for the fluorescence change obtained. The 2D structures of AOLE constituents were drawn using ChemDraw Ultra (version 8.0, Cambridgesoft Com., USA). Chem3D Ultra was used to convert 2D structure into 3D and the energy was minimized using the semi-empirical AM1 method. Minimized energy to minimum RMS gradient of 0.100 was set in each iteration. All structures were saved as PDB file format for input to ADT. All AOLE constituents chemical structures were then saved in PDBQT file format to carry out docking in ADT. The crystal structures of bee-venom phospholipase A2 and the cobra-venom phospholipase A2 in a complex with a transition-state analogue were downloaded from the Protein Data Bank. In addition, the crystal structure of melittin was downloaded from the Protein Data Bank. The molecular docking of AOLE contents with both snake and bee phospholipase A2 and melittin were accomplished by AutoDock 4.2 software from the Scripps Research Institute (TSRI). Firstly, the polar hydrogen atoms were added into phospholipase A2 and melittin molecules. Then, the partial atomic charges of phospholipase A2 and melittin and AOLE contents molecules were calculated using Kollman methods [19]. In the process of molecular docking, the grid maps of dimensions (62Å X 62Å X 62Å) with a gridpoint spacing of 0.376 Å and the grid box centered. The number of genetic algorithm runs and the number of evaluations was set to 100. All other parameters were default settings. Cluster analysis was performed on the results of docking by using a Root Mean Square (RMS) tolerance of 2.0 Å, dependent on the binding free energy. Lastly, the dominating configuration of the binding complex of AOLE contents and phospholipase A2 and melittin proteins with minimum energy of binding determined.
Results and Discussion
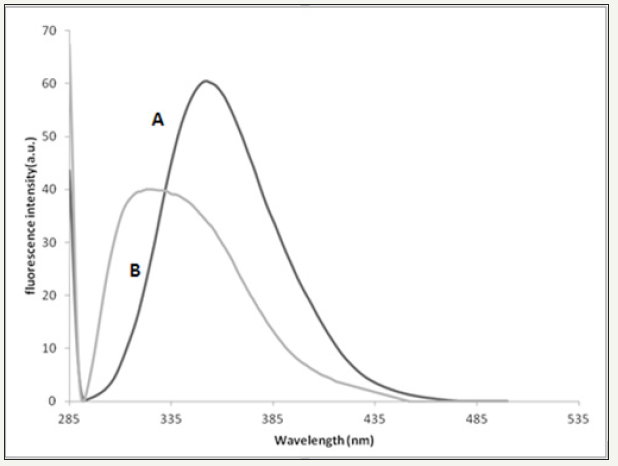
All snake venoms of Cerastes cerastes, Macrovipera deserti and Echis pyramidum behave similarly with AOLE using fluorescence spectroscopy; only the fluorescence results of Cerastes cerastes are shown in this work. The fluorescence spectrum shows a decrease of fluorescence intensity (Figure 1) of the snake venom due to addition of 100μl of AOLE which could be related to various processes. It is well known that a decrease in fluorescence intensity can be caused by a range of molecular interactions such as molecular rearrangements, excited-state reactions, ground state complex formation, collisional quenching or energy transfer. The decrease in fluorescence emission intensity as shown in Figure 1 was accompanied by a red shift of 7 nm of the maximum emission (343nm to 350nm) which may indicate that Trp residues buried in a hydrophobic environment have moved into a relatively polar environment consistent with earlier reports [20]. The decrease in fluorescence emission intensity (Figure 2) was accompanied by a blue shift of 24 nm of the maximum emission (352 to 328nm) and this may indicate that binding of AOLE components may have accomplished a conformational change that moves Trp into a relatively more hydrophobic region.
figure 1: (A) Fluorescence perturbation of snake venom by addition of AOLE. Plot of fluorescence emission of snake venom (Cerastes cerastes) (24.6μg/ml) vs wavelength from 288-540 nm using excitation of λ280 nm in 0.01 M Tris, 0.1 M NaCl at pH 7.4. (B)Plot of fluorescence emission of snake venom Cerastes cerastes (24.6μg/ml) and 100 μl AOLE (5g/15ml) vs wavelength from 288-540 nm using excitation of λ280 nm in 0.01 M Tris, 0.1 M NaCl at pH 7.4. Spectra were corrected for small background fluorescence contributions from the buffer solution and were scaled to visualize the shift.
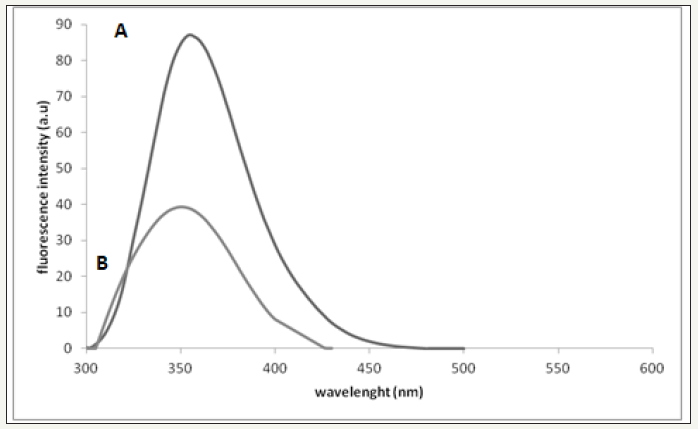
figure 2: (A)Fluorescence perturbation of bee venom by addition of AOLE. Plot of fluorescence emission of bee venom (26μg/ ml, each 1mg of dry venom contains 6.6μg/ml protein) vs wavelength from 288 - 540 nm using excitation of λ280 nm in 0.01 M Tris, 0.1 M NaCl at pH 7.4. (B)Plot of fluorescence emission of bee venom (26μg/ml) and 100μl AOLE (5g/15ml) vswavelength from 288-540nm using excitation of λ280 nm in 0.01M Tris, 0.1M NaCl at pH 7.4. Spectra were corrected for small background fluorescence contributions from buffer solution and were scaled to visualize the shift.
Similar results report an increase in intrinsic tryptophan fluorescence and a blue shift of the maximum emission wavelength upon addition of MgATP to ArsA ATPase, indicating movement of Trp-159 into a relatively less polar environment [21]. The fluorescence of Phospholipases A2 if excited at 280nm is mainly due to the presence of a single Tryptophan residue (Trp3) which is located on the surface of the enzyme molecule exposed to the environment [22].
The interaction between AOLE and the snake or bee venoms is not a concentration-dependent quenching of the fluorescence of the venom. Hence it is difficult to study fluorescence quenching which usually originates from collision between the fluorophore and the quencher (dynamic quenching) and/or is due to complex formation between the fluorophore and the quencher (static quenching) [23]. In addition, due to the red or blue shift of the fluorescence spectra, as seen in Figure 1 & 2, it is difficult to determine the mechanism of interaction between fluorescence of both venoms and AOLE. Therefore, analysis by the Stern-Volmer equation to determine the quenching constant was not possible.
Solvent polarity and the local environment have profound effects on the spectral emission properties of fluorophores. The effect of solvent polarity is an important determinant of the Stokes shift, which is one of the earliest observations in fluorescence and was seen clearly in our experiments. Emission spectra are easily measured, resulting in numerous publications on emission spectra of fluorophores in different solvents and bound to proteins, membranes, DNA or RNA. One general use of solvent effects is to determine the polarity of the probe binding site at the macromolecule. This is accomplished by comparison of emission spectra and/or quantum yields if the fluorophore is bound to the macromolecule or dissolved in solvents of different polarity. However, there are many additional instances where solvent effects are used. We used them when the fluorescent bee and snake venoms bind to any component of AOLE. The binding was accompanied by a spectral shift or change in quantum yield due to the different environment for the bound fluorescence component. The fluorescence spectral changes can be used to measure the extent of binding between the fluorescent protein and the AOLE components. The effects of the environment on fluorescence spectra and quantum yields are complex and are due to several factors including: solvent polarity, viscosity, rate of solvent relaxation, probe conformational changes, rigidity of the local environment, internal charge transfer, proton transfer, excited state reactions, probe-probe interactions or changes in radiative and non-radiative decay rates. These multiple effects may offer chances to probe the local environment surrounding a fluorophore. However, environmental effects are usually complex and even solvent polarity cannot be described using a single theory. The Lippert-Mataga equation partially explains the effect of solvent polarity, but does not account for other effects such as hydrogen bonding to the fluorophore or internal charge transfer that depends on solvent polarity [24,25]. Emission spectra are seen to shift considerably to longer or shorter wavelengths as the solvent polarity changes upon addition of AOLE. The sensitivity to solvent polarity could be related to electron-donating or -accepting groups on tryptophan. In addition, the hydroxyl group of polyphenols in AOLE serves as electron donor and the carbonyl group on tryptophan as electron acceptors. This condition might help to understand the observed fluorescence changes in snake and bee venoms dissolved in AOLE. The high sensitivity to the solvent is due to a charge shift away from the hydroxyl groups in the excited state towards the electron acceptor. This results in a large dipole moment in the excited state. The dipole moment interacts with the polar solvent molecules to reduce the energy of the excited state and hence decreases fluorescence intensity.
(Figure 1A) Fluorescence perturbation of snake venom by addition of AOLE. Plot of fluorescence emission of snake venom Cerastes cerastes (24.6μg/ml) vs wavelength from 288-540nm using excitation of λ280 nm in 0.01 M Tris, 0.1 M NaCl at pH 7.4. (B) Plot of fluorescence emission of snake venom Cerastes cerastes (24.6μg/ml) and 100μl AOLE (5g/15ml) vs wavelength from 288- 540nm using excitation of λ280 nm in 0.01M Tris, 0.1M NaCl at pH 7.4. Spectra were corrected for small background fluorescence contributions from the buffer solution and were scaled to visualize the shift.
(Figure 2A) Fluorescence perturbation of bee venom by addition of AOLE. Plot of fluorescence emission of bee venom (26μg/ml, each 1mg of dry venom contains 6.6μg/ml protein) vs wavelength from 288 - 540 nm using excitation of λ280 nm in 0.01 M Tris, 0.1 M NaCl at pH 7.4. (Figure 2B) Plot of fluorescence emission of bee venom (26μg/ml) and 100μl AOLE (5g/15ml) vs wavelength from 288-540nm using excitation of λ280nm in 0.01 M Tris, 0.1 M NaCl at pH 7.4. Spectra were corrected for small background fluorescence contributions from buffer solution and were scaled to visualize the shift.
Tryptophan fluorescence-bee venom
Bee venom PLA2 contains two tryptophan residues [26,27] one near the N-terminus, Trp-8 and one near the C-terminus, Trp-128. In addition it has 8 tyrosine residues distributed throughout the protein sequence. The fluorescence emission spectrum of the native protein (λmax = 352nm) showed that the Trp residues were in a moderately hydrophobic environment [26,27]. When the enzyme was treated with AOLE the fluorescence emission first decreased sharply and then declined progressively, undergoing a small, but detectable blue shift (Figure 2). The simplest interpretation of these results is that the highly hydrophobic contents of the AOLE binds to the enzyme very rapidly, perturbing the environment of one of the tryptophan residues, and then undergoes a relatively slow reaction in which the AOLE components were transferred to an acceptor residue which leads to further perturbation of the tryptophan environment. In addition, melittin is the major component of bee venom and it is crystal structure has been solved, which indicated a tetrameric helix-bend-helix structure with two helical segments, positioned between amino acid residues 1-10 and 13-26 and the bend region within residues 11-12 [28]. The blue-shift obtained could be also interpreted as the Trp residue of melittin moving into an environment with a lower dielectric constant and this is consistent with the same results obtained by Burstein et al. [29].
Molecular docking analysis
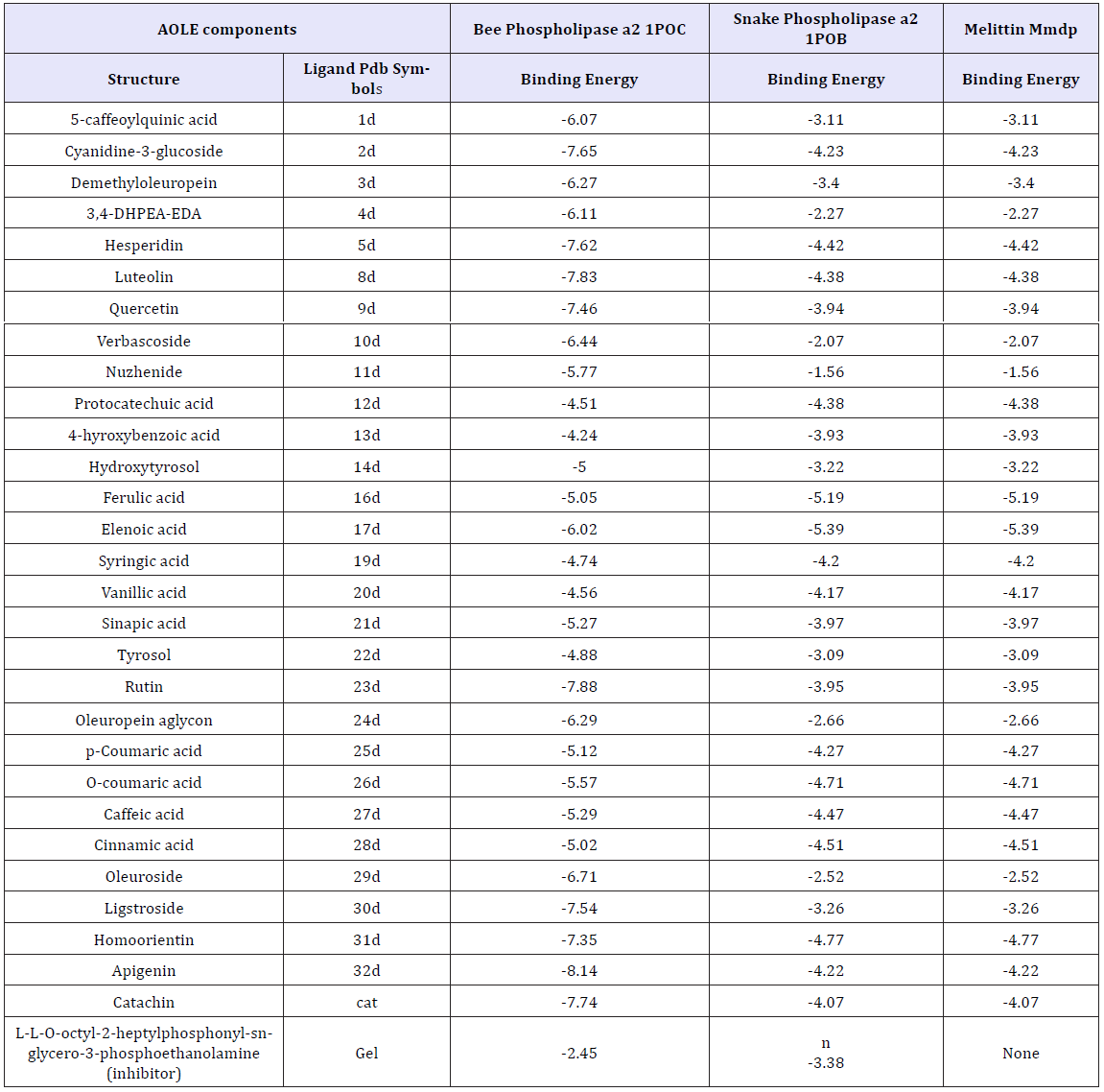
Table 1 shows the energies of binding of the AOLE contents with bee and venom phospholipase A2 and melittin obtained by using molecular docking strategy. aΔG is the free binding energy change in the binding process (Kcal/mole)Molecular docking of AOLE contents with phospholipase A2 and melittin were performed using AutoDock 4.2 to investigate the binding mode of AOLE contents with phospholipase A2 and melittin and to obtain information about the interaction forces between AOLE contents and phospholipase A2 and melittin. AOLE which consists of flexible molecules was docked to two types of rigid phospholipase A2 and melittin proteins to obtain the preferential binding site(s) of AOLE to phospholipase A2 and melittin [30-32]. The molecular docking results are shown in Table 1. modeling study indicates substantial interactions between AOLE contents and bee and snake PLA2. The docking binding energies are shown in Table 1. For example, apigenin (an AOLE component) in case of snake PLA2 was able to form hydrogen bonds with the residues Gly29, Leu249, Tyr27, Asp48 and Tyr63 of the enzyme, pi-pi stacking with the residue Phe5, Phe5,His47 and His47 and Pi-alkyl interaction with the residue Leu2. In addition, the molecular docking results indicate that other amino acid residues are involved in the interactions with AOLE contents. Similar binding energies of docking were obtained using AOLE with bee PLA2 as shown in Table 1. For example the AOLE component apigenin in case of bee PLA2 was able to form hydrogen bonds with the residues Phe82, His34 (two bonds) and Asp35 and Tyr57 of the enzyme, as well as pi-pi stacking with the residue Tyr87, Pi-alkyl interaction with the residue Cys8, Met86 and pi-sigma interaction with the residue Val83. The interactions of AOLE components (which were mainly polyphenolic components) obtained are in consistency with previous studies reporting that polyphenolic secondary metabolites are able to inhibit PLA2 [33]. For example quercetin, kaempferol, galangin, naringenin, artemetin and other flavonoids can inhibit toxins from snake venom. They found that flavonoids usually exert their inhibitory effect through hydrophobic interactions with A and B rings and aromatic or hydrophobic amino acid residues in the protein [34-36]. In addition, another research group found that Ar-turmerone which is a phenolic compound isolated from the Curcuma longa (Zingiberaceae) plant has a strong inhibitory effect against hemorrhage and lethality caused by B. jararaca and C. d. terrificus snake venom. The effect was attributed to the interaction with PLA2 [37]. It has been also reported that 4-nerolidylcatechol (a hydroxylated phenolic compound) - an extract from Piper umbellatum and P. peltatum (Piperaceae) - is able to inhibit the myotoxic activities of PLA2s [38] and is able to interact and inhibit the function of PLA2s. Araya and Lomonte [39] found that caffeic acid which is one of the components of AOLE, can interact with proteins via hydrogen bonds, inhibiting enzyme function and acting as antidote. They found that strong interactions may cause conformational changes in the protein structure [39,40]. The interaction of caffeic acid with snake venoms were confirmed by Shimabuku and collaborators [41] who crystallized PrTX-I (basic Lys49-PLA2 from B. pirajai snake venom) in the presence of the inhibitor caffeic acid. The electron-density map clearly signifies the presence of three caffeic acid molecules interacting with the C-terminus of the protein. The changes of fluorescence emission intensity and fluorescence shifts (blue and red) in which the formation of the system was formed by sequential addition of aliquots of Tris buffer, snake and bee venoms and finally AOLE (Figure 1& 2) confirms interaction obtained by the modeling studies.
The docking binding energies obtained from AOLE interacting with bee-melitten as shown in Table 1 was due to the formation of hydrogen bonds with the residues Lys21 (three bonds), Ser18, and Leu9 of the enzyme, and pi-sigma interaction with the residue Ile17. In addition, the molecular docking results showed that the residues Ile17 (two bonds), Pro14 (two bonds), and Ile13 are involved in the pi-alkyl interactions with apigenin. Results provided by Chatterjee et al. [42] reported that melittin interacts with ganglioside GM1 which is consistent with our findings and were confirmed by different techniques including steady-state fluorescence, molecular modeling and proton NMR spectroscopy [42]. They found that in the presence of GM1 the emission maximum of melittin was blue shifted and fluorescence quenching efficiencies of iodide and acrylamide were substantially reduced, indicating shielding of tryptophan of melittin from the aqueous environment. In their study with the aid of molecular modeling they confirm a melittin- GM1 complex with a N-terminal hydrophobic stretch of melittin connected through the ceramide tail and C-terminal hydrophilic end of melittin encompassing constructive electrostatic interaction with the carbohydrate head group of GM1 [42].
Table 1: Various energies in the binding process ofAOLE contents with bee and venom phospholipase A2 and melittin obtained from molecular docking. The unit of all energies was kJ/mol.
Conclusion
Changes in fluorescence spectra indicate that the occupation of AOLE component binding sites cause conformational modifications of Phospholipase A2 and melittin. The observed red-shift in the emission maximum suggests that the conformational change increases the exposure of at least one tryptophan residue to the solvent. As expected, the effect is immediate when induced by addition of AOLE components. Of the two Trp residues of bee PLA2, Trp-8 is located at a relatively internal hydrophobic environment near the active site, whilst Trp-128 is in a less compact region of the protein with a lower density of hydrophobic residues. Hence, Trp-8 is the more likely source of the observed changes. It is of interest to note that bee PLA2 interacting with AOLE components gives significantly larger fluorescence changes compared to the snake venom suggesting that the two adjacent Trp residues could be involved. Evidence presented here shows that activation of PLA2 is based on conformational modifications associated with occupation of a hydrophobic site on PLA2. In further studies using the fluorescence technique we will investigate the relationship between structural changes in the bee and snake venoms.
References
- de Roodt AR, Dolab JA, Dokmetjian JC, Litwin S, Segre L, et al. (2000) A comparison of different methods to assess the hemorrhagic activity of Bothrops venoms. Toxicon 38(6): 865-873.
- . Mohamed AH, Ibrahim FK, Fattah MM, Ramandan M, Darwish M (1977) Toxic fractions of Cerastescerastes venom. Jpn J Med Sci Biol 30(4): 205- 207.
- Siddiqi AR, Shafqat J, Zaidi ZH, Jornvall H (1991) Characterization of phospholipase A2 from the venom of Horned viper (Cerastes cerastes). FEBS Lett 278(1): 14-16.
- Gasanov SE, Gasanov NE, Rael ED (1995) Phospholipase A2 and cobra venom cytotoxin Vc5 interactions and membrane structure. Gen PhysiolBiophys 14(2): 107-123.
- Alzahaby M, Rowan EG, Young LC, al Zahaby AS, Abu-Sinna G, et al. (1995) Some pharmacological studies on the effects of Cerastesvipera (Sahara sand viper) snake venom. Toxicon 33(10): 1299-1311.
- Cavalcante WLG, Noronha-Matos JB, Timoteo MA, Fontes MRM, Gallacci M, et al. (2017) Neuromuscular paralysis by the basic phospholipase A2 subunit of crotoxin from Crotalusdurissus terrificus snake venom needs its acid chaperone to concurrently inhibit acetylcholine release and produce muscle blockage. ToxicolAppl Pharmacol 334: 8-17.
- Pradhan S, Kumar S, Singh D, Sood RC, Sehgal R (2007) Development of Passive Haemagglutination (PHA) and Haemagglutination Inhibition (HAI) technique for potency estimation of Cobra Antisnake Venom Serum (ASVS)Biologicals 35(3): 155-160.
- Theakston RD, Reid HA (1983) Development of simple standard assay procedures for the characterization of snake venom. Bull World Health Organ 61(6): 949-956.
- Gasparello-Clemente E, Silveira PF (2002) Fluorometric assay using naphthylamide substrates for assessing novel venom peptidase activities. Toxicon 40(11): 1617-1626.
- Kebir-Chelghoum H, Laraba-Djebari F (2017) Cytotoxicity of Cerastescerastes snake venom: Involvement of imbalanced redox status. Acta Trop 173: 116-124.
- Reid HA (1975) Epidemiology of sea-snake bites. J Trop Med Hyg78(5): 106-113.
- Fonger GC, Hakkinen PJ, Publicker S (2015) Venoms and antivenoms: North American poisonous scorpion, snake, and spider information is now in the National Library of Medicine’s Hazardous Substances Data Bank. Clin Toxicol (Phila) 53(1): 74.
- Fischer FG, Neumann WP (1961) [The venom of the honeybee. III. On the chemical knowledge of the principle active constituent (melittin)]. Biochem Z335: 51-61.
- Hartter P (1977) [Basic peptides in bee venom, III. Synthesis of peptide fragments from the sequence of the mast-cell-degranulating peptide (author’s transl)]. Hoppe Seylers Z Physiol Chem 358(3): 331-337.
- Shipolini RA, Callewaert GL, Cottrell RC, Doonan S, Vernon CA, et al. (1971) Phospholipase A from bee venom. Eur J Biochem 20(4): 459-468.
- Orlov BN, Omarov S, Gelashvili DB, Korneva NV, Asafova NN (1978) [Chemistry and pharmacology of bee venom (a review of the literature)]. FarmakolToksikol41(3): 358-369.
- Kamceva T, Flemmig J, Damnjanovic B, Arnhold J, Mijatovic A, et al. (2011) Inhibitory effect of platinum and ruthenium bipyridyl complexes on porcine pancreatic phospholipase A2. Metallomics 3(10): 1056-1063.
- Markwell MA, Haas SM, Bieber LL, Tolbert NE (1978) A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal Biochem 87(1): 206-210
- Tiwari R, MahasenanK, Pavlovicz R, Li C, Tjarks W (2009) Carborane clusters in computational drug design: a comparative docking evaluation using AutoDock, FlexX, Glide, and Surflex. J Chem Inf Model 49(6): 1581- 1589.
- Kenoth R, Simanshu DK, Kamlekar RK, Pike HM, Molotkovsky JG, et al. (2010) Structural determination and tryptophan fluorescence of heterokaryon incompatibility C2 protein (HET-C2), a fungal glycolipid transfer protein (GLTP), provide novel insights into glycolipid specificity and membrane interaction by the GLTP fold. J Biol Chem285(17): 13066- 13078.
- Zhou T, Rosen BP (1997) Tryptophan fluorescence reports nucleotideinduced conformational changes in a domain of the ArsA ATPase. J Biol Chem272(32): 19731-19737.
- Cunningham TJ, Yao L, Lucena A (2008) Product inhibition of secreted phospholipase A2 may explain lysophosphatidylcholines’ unexpected therapeutic properties. J Inflamm 5: 17.
- Khot MS, Bhattar SL, Kolekar GB, Patil SR (2010) Spectrofluorimetric determination of 5-fluorouracil by fluorescence quenching of 9-anthracenecarboxylic acid. Spectrochim Acta A Mol BiomolSpectrosc 77(1): 82-86.
- Snitsarev V, Young MN, Miller RM, Rotella DP (2013) The Spectral Properties of (-)-Epigallocatechin 3-O-Gallate (EGCG) Fluorescence in Different Solvents: Dependence on Solvent Polarity. PLoS One 8(11): e79834.
- Gong Y, Guo X, Wang S, Su H, Xia A, et al. (2007) Photophysical properties of photoactive molecules with conjugated push-pull structures. J Phys Chem A 111(26): 5806-5812.
- KuchlerK, Gmachl M, Sippl MJ, Kreil G (1989) Analysis of the cDNA for phospholipase A2 from honeybee venom glands. The deduced amino acid sequence reveals homology to the corresponding vertebrate enzymes. Eur J Biochem 184(1): 249-254.
- Chang ZY, Nygaard P, Chinault AC, Kellems RE (1991) Deduced amino acid sequence of Escherichia coli adenosine deaminase reveals evolutionarily conserved amino acid residues: implications for catalytic function. Biochemistry 30(8): 2273-2280.
- Quay SC, Condie CC (1983) Conformational studies of aqueous melittin: thermodynamic parameters of the monomer-tetramer self-association reaction. Biochemistry 22(3): 695-700.
- Burstein EA, Vedenkina NS, Ivkova MN (1973) Fluorescence and the location of tryptophan residues in protein molecules. PhotochemPhotobiol 18(4): 263-279.
- Scott DL, Otwinowski Z, Gelb MH, Sigler PB (1990) Crystal structure of bee-venom phospholipase A2 in a complex with a transition-state analogue. Science250(4987):1563-1566.
- White SP, Scott DL, Otwinowski Z, Gelb MH, Sigler PB (1990) Crystal structure of cobra-venom phospholipase A2 in a complex with a transition-state analogue. Science 250(4987): 1560-1563.
- Schlamadinger DE, Wang Y, McCammon JA, Kim JE (2012) Spectroscopic and computational study of melittin, cecropin A, and the hybrid peptide CM15. J Phys Chem B 116(35): 10600-10608.
- Cotrim CA, de Oliveira SC, Diz Filho EB, Fonseca FV, Baldissera L, et al. (2011) Quercetin as an inhibitor of snake venom secretory phospholipase A2. Chem Biol Interact 189(1-2) :9-16.
- Ticli FK, Hage LI, Cambraia RS, Pereira PS, Magro AJ, et al. (2005) Rosmarinic acid, a new snake venom phospholipase A2 inhibitor from Cordia verbenacea (Boraginaceae): antiserum action potentiation and molecular interaction. Toxicon 46(3): 318-327.
- Soares AM, Ticli FK, Marcussi S, Lourenco MV, Januario AH, et al. (2005) Medicinal plants with inhibitory properties against snake venoms. Curr Med Chem12(22): 2625-2641.
- Lattig J, Bohl M, Fischer P, Tischer S, Tietbohl C, et al. (2007) Mechanism of inhibition of human secretory phospholipase A2 by flavonoids: rationale for lead design. J Comput Aided Mol Des 21(8): 473-483.
- Ferreira LA, Henriques OB, Andreoni AA, Vital GR, Campos MM, et al. (1992) Antivenom and biological effects of ar-turmerone isolated from Curcuma longa (Zingiberaceae). Toxicon30(10): 1211-1218.
- Nunez V, Castro V, Murillo R, Ponce-Soto LA, Merfort I, et al. (2005) Inhibitory effects of Piper umbellatum and Piper peltatum extracts towards myotoxic phospholipases A2 from Bothrops snake venoms: isolation of 4-nerolidylcatechol as active principle. Phytochemistry 66(9): 1017-1025.
- Araya C, Lomonte B (2007) Antitumor effects of cationic synthetic peptides derived from Lys49 phospholipase A2 homologues of snake venoms. Cell Biol Int 31(3): 263-268.
- Lomonte B, Angulo Y, Moreno E (2010) Synthetic peptides derived from the C-terminal region of Lys49 phospholipase A2 homologues from viperidae snake venoms: biomimetic activities and potential applications. Curr Pharm Des 16(28): 3224-3230.
- Shimabuku PS, Fernandes CA, Magro AJ, Costa TR, Soares AM, et al. (2011) Crystallization and preliminary X-ray diffraction analysis of a Lys49-phospholipase A2 complexed with caffeic acid, a molecule with inhibitory properties against snake venoms. Acta Crystallogr Sect F Struct BiolCrystCommun 67(Pt 2): 249-252.
- Chatterjee C, Mukhopadhyay C (2002) Melittin-GM1 interaction: a model for a side-by-side complex. BiochemBiophys Res Commun292(2): 579-585.
© 2018 Abdul M Gbaj. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)













.bmp)
.jpg)
.png)
.jpg)














.png)

.png)



.png)






