- Submissions

Full Text
Cohesive Journal of Microbiology & Infectious Disease
New Haplotype of Pseudocohnilembus (Ciliophora: Cuticociliate) Identified from a Rock Dove Feces in Algeria
Asma Guilane1, Tahar Kernif2, Fadila Tazerouti1, Rezak Drali3 and Amina Boutellis1*
1Biodiversity and Environment Laboratory, Houari Boumediene University of Science and Technology, Algeria
2Laboratory of Parasite Eco-Epidemiology and Population Genetics, Pasteur Institute of Algeria, Algeria
3Genomics Platform-Bioinformatics, Pasteur Institute of Algeria, Algeria
*Corresponding author: Amina Boutellis, LBEIG, Faculty of Biological Sciences, Houari Boumediene University of Science and Technology, Algiers
Submission: January 12, 2024; Published: February 01, 2024

ISSN 2594-0190 Volume7 issues2
Abstract
Scuticociliates are facultative parasitic ciliates of the subclass Scuticociliatia Small, 1967 which cause scuticociliatosis, one of the most important parasitological problems in marine aquaculture worldwide. In this study, we report, for the first time, the presence of Pseudocohnilembus sp. in Algeria. The unexpected discovery of Scuticociliate DNA during our research on protozoan parasites in bird faeces, led to the parasite’s molecular and phylogenic characterization. BLAST analysis of the partial small subunit ribosomal DNA sequence (SSU rDNA) showed sequence similarity value of 99.64% with an uncultured Pseudocohnilembus clone. The alignment of this sequence with other Pseudocohnilembus sequences gives a separated sub-clade among the same uncultured Pseudocohnilembus sequences isolated from stool. The distance between our Pseudocohnilembus sp. and the uncultured Pseudocohnilembus sequences isolated from stool does not exceed 1% when computed using Kimura 2-parameter model, however, it greatly exceeds 1.5% with the other species included in the dataset. These distances, and the phylogenic tree, clearly indicates that Pseudocohnilembus sp. isolated from different animal stools corresponds to a new species, and our present sequence of Pseudocohnilembus sp. forms an original Algerian haplotype.
Keywords: Animal feces; Birds; Genotype; Protozoa; SSU rDNA; Scuticociliates
Introduction
Ciliated protozoa are single-cell eukaryotes with diverse morphologies and extensive distributions [1]. Scuticociliatia, one of six subclasses of the class Oligohymenophorea, are commonly found in ecosystems worldwide [1,2]. They are known to rapidly invade and establish colonization of the marine host organs like gills, skin, brain, heart, muscles including visceral organs, and intestine [3,4]. Scuticociliatosis is highly histophagous and destroys infected tissues [5]. Outbreaks of scuticociliates cause mass mortalities in fishes, and has been reported in olive flounder Paralichthys olivaceus, rainbow trout Oncorhynchus mykiss, southern bluefin tuna Thunnus maccoyii, turbot Scophthalmus maximus, seabass Dicentrachus labrax, and silver pomfret Pampus argenteus [3]. The genus Pseudocohnilembus Evans and Thompson (1964) was originally erected for free-living marine ciliate [6,7]. However, since isolation of P. persalinus from diseased olive flounder in Korea by Kim et al. [8], it has become recognized as an important facultative parasite causing serious economic losses in marine aquaculture worldwide. Genome analysis showed that P. persalinus has acquired many unique prokaryote-derived genes that potentially contribute to the virulence of this organism, including cell adhesion, hemolysis, and heme utilization genes [2]. Moreover, P. persalinus is an ideal model organism for a wide variety of biological research [9] and is considered to be a good microorganism to evaluate environmental risks by applying nanomaterial monitoring and bioremediation [10]. The SSU rDNA sequence analysis approach is a universally applicable tool to identify scuticociliates because morphological analysis, based upon qualitative staining techniques may obscure subtle variations, and lead to misidentification [3]. This study was first aimed to screen intestinal protozoan parasite present in birds, and the fortuitous discovery of a Scuticociliate DNA led us to a molecular and phylogenic characterization of the parasite.
Materials and Methods
Collection of animal samples
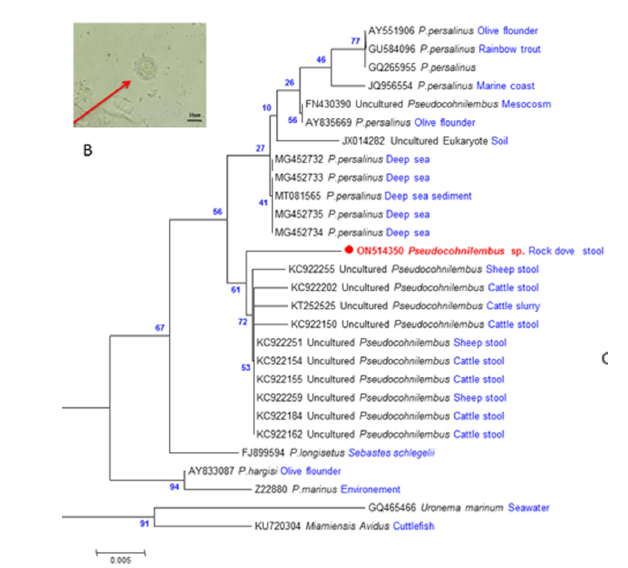
This study was conducted in accordance with the World Animal Health Organization [11] guiding principles on animal welfare included in the OIE Terrestrial Animal Health Code. Fresh fecal samples from different bird species collected from Algiers, North Algeria (36° 45’ 9.00” N and 3° 02’ 31.09” E), were screened for gastrointestinal parasites using the zinc sulphate flotation method, followed by microscopy [12]. Among the analyzed samples, one belonging to a rock dove Columba livia Gmelin, 1789, presented granular forms similar to Blastocystis sp (Figure 1).
Figure 1:A. Microscopy of the granular form of Pseudocohnilembus species observed on the coprological examination. B. Molecular Phylogenetic analysis of the SSU rDNA of Pseudocohnilembus sequences are designated by the GenBank access number and the name of their hosts or their isolation source. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 28 nucleotide sequences. All positions containing gaps and missing data were eliminated. There are a total of 284 positions in the final dataset. Evolutionary analyses were conducted by ML method in MEGA7.
Molecular studies
The selected sample was conserved in 70% ethanol, and then washed with milliQ water prior to the DNA extraction method from fresh stool. DNA was extracted using the QIAGEN DNA Tissue Kit (Qiagen, Courtaboeuf, France) according to the manufacturer’s protocol, with the modifications previously specified [13,14]. The presence of the so-called Blastocystis in DNA samples was determined using PCR targeting a fragment of the SSU rDNA gene using previously described primers [15]. PCR reactions were performed in 25μl reaction mixtures containing 2.5μl of 10× buffer, 2.5mM of MgCl2, 200μM of each Deoxy-Ribonucleoside Triphosphate (dNTPs) mixture, 0.2μM of each primer (Eurogentec, Belgium), 0.025U/μl of Taq DNA polymerase Hot Star (Qiagen, Courtaboeuf, France), and 5μl of genomic DNA using the following thermocycling conditions: 95 °C for 15min, followed by 95 °C for 30s, 54 °C for 30s, and 72 °C for 1min for 35 cycles, and a final extension 72 °C for 5min. PCR amplification result was then verified by electrophoresis migration of PCR product on a 1.5% agarose gel using Ethidium bromide. Nucleo Fast 96PCR Plates (Macherey-Nagel EURL, France) and BigDye Terminator version 1.1 cycle sequencing-ready reaction mix (Applied Biosystems, Foster City, CA) were then used to purify one positive PCR product to be sequenced directly in both directions with the same primers used in the PCR amplification. The ABI 3100 automated sequencer (Applied Biosystems) resolved the sequenced products. The obtained sequence was edited with Chromas Pro software (Technelysium PTY, Australia), compared to the GenBank database using BLAST, and deposited in GenBank under the accession number ON514350. The phylogeny reconstruction and genetic distances were inferred using the MEGA7 software [16]. Haplotype Diversity (Hd) and nucleotide diversity (π) were calculated using DNA Sequence Polymorphism (DNASP 5.10.01) [17].
Results
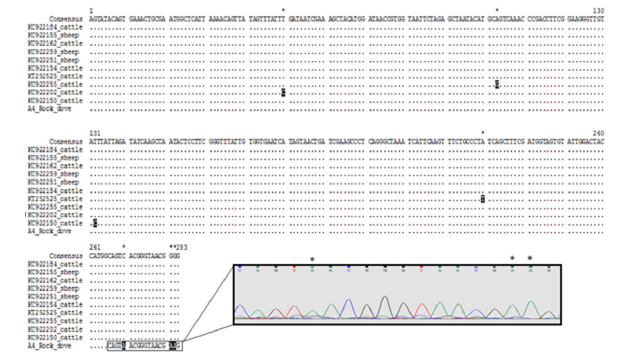
The provisional identification of protozoan species using morphological characteristics was challenged by DNA barcoding approach. In the present study, BLAST analysis of the partial SSU rDNA (284bp) sequence of the protozoan showed 99.64% sequence similarity with an uncultured Pseudocohnilembus clone (KC922259) isolated from a sheep stool in China. The SSU rDNA partial sequence of Pseudocohnilembus sp. was aligned with 25 other Pseudocohnilembus sequences (uncultured Pseudocohnilembus, P. persalinus, P. longisetus, P. hargisi and P. marinus) (Table 1). Sequences of Miamiensis Avidus and Uronema marinum were used as the out-group. The trees inferred using the Neighbor-Joining (NJ) and Maximum Likelihood (ML) led to similar topologies and thus only the ML tree is shown (Figure 1). For the latter, the best model, estimated by MEGA7, was the Tamura 3-parameter with a discrete Gamma Distribution (G) [16]. All Pseudocohnilembus species formed a monophyletic clade separated from the out groups (91% bootstrap). The Pseudocohnilembus clade exhibits 3 sub-clades: P. hargisi and P. marinus clustered separately together (94% bootstrap), P. longisetus forms a separate cluster (67% bootstrap), and the Pseudocohnilembus sequence of the present study is composed with other uncultured Pseudocohnilembus sequences, a sister group with P. persalinus clade (56% bootstrap). Distances computed using Kimura 2-parameter distance between our Pseudocohnilembus sp. sequence and uncultured Pseudocohnilembus sequences isolated from stool corresponds to 1%, and distances between our sequence and the other species ranged between 2-3% (P. persalinus), 3-4% (remaining Pseudocohnilembus species) and 5-6% (out groups) (Figure 2). Considering that the interspecific genetic distances of ciliates are more than 1.5% [18], these distances, and the tree, clearly indicate that Pseudocohnilembus sp. isolated from different animal stools corresponds to a new species. Multiple sequence alignment of the uncultured Pseudocohnilembus species isolated from stool revealed limited heterogeneity among the 11 sequences (nucleotide diversity π = 0.00450), in the form of 7 singleton variable sites leading to the exhibition of four different haplotypes with a rate of gene diversity Hd corresponding to 0.727 (Figure 3). Our present sequence of Pseudocohnilembus sp. forms an original algerian haplotype supported by 61% bootstrap value (Figure 1).
Table 1:Species of Pseudocohnilembus used in the present molecular study. All species belong to the Scuticociliatia.

Figure 2:Estimate of evolutionary divergence between sequences. There were a total of 284 positions in the dataset. Distances are Kimura 2-parameter distances, shown as percentages.

Figure 3:Multiple sequence alignment of the uncultured Pseudocohnilembus species isolated from stool.
Discussion
In the present study, a new species of Pseudocohnilembus associated to animal stools was obtained by sequencing the partial SSU rDNA. The sequence is clean and much perfectly with Blastocystis primers designed by Poirier et al. [15], as well as the negative control remained negative all along amplification and sequencing. In literature, several Scuticociliate species have already been reported in farmed marine fish and deep sea [8,19]. However, only two publications report the presence of eukaryotic Scuticociliate out of seawater, especially in soil [20] and in a farm anaerobic reactor inoculated with cattle slurry [21]. Here, the obtained Pseudocohnilembus sp. sequence was isolated from a stool sample of a rock dove in Algiers. The phylogenic characterization shows that our sequence clustered with uncultured Pseudocohnilembus sequences isolated from cattle and sheep stool from China (https:// www.ncbi.nlm.nih.gov/nuccore/KC922255 unpublished data). The uncultured eukaryote identified in the French anaerobic reactor inoculated with stale and cold cattle slurry also clustered on the same clade forming a monophyletic stool Pseudocohnilembus clade. The sister clade corresponds to Pseudocohnilembus persalinus, which has been recognized as a common facultative pathogen [7]. In recent years, there have been many reports of fatal outbreaks of infection in marine fish by several Scuticociliate species including Pseudocohnilembus persalinus [2]. The gene sequencing of microeukaryotes isolated from the Yellow River delta in China revealed a higher diversity of ciliate assemblages including P. persalinus in the high salinity soil adjacent to the sea [20], and Pseudocohnilembus have been tested as an appropriate indicator of high salinity levels in soil [22].
Rock dove or pigeon belong to order Columbiformes, and family Columbidae have been associated with humans for a long time now. They are raised as ornamental birds, companion animals, meat production or laboratory specimens [23]. Dove’s parasitic infections are widespread, especially coccidiosis [24]. The causative agent of this disease is in the genus Eimeria: E. columbarum, E. labbeana, and E. columbae [23]. Another important disease of doves is Trichomoniasis [25]. Doves were considered as the primary reservoir of Trichomonas gallinae [26], otherwise, since the report of Colpoda steinii in oral swabs from mourning doves (Zenaida macroura) by Toepfer Jr [27] non ciliate was reported in pigeons.
Conclusion
In conclusion, this is the first report of Pseudocohnilembus sp. in Algeria. The molecular identification of the parasite in a stool sample of a rock dove leads us to speculate that it may have come from a different water source in the city of Algiers, in particular the Mediterranean Sea. However, the genetic profile of this organism is different from other marine origin species. As a result, we believe that a larger number and variety of birds and other animals should be tested in the future in this area. Moreover, further studies using specific primers of Scuticociliates are needed to confirm our study and to determine if this genetic profile of Pseudocohnilembus sp is original and specific to birds in Algeria.
Acknowledgment
We gratefully thank Dr. Poonam SHARMA, Oklahoma State University, United States of America for her diligent proofreading of this article. We also thank lillia lidia BELALOUI and Hedia BENMOUHOUB for their contributions. This research was supported by the ‘Direction Générale de la Recherche Scientifique et du Développement Technologique DGRSDT and Pasteur Institute of Algeria.
References
- Pan X, Yi ZJ, Li J, Ma H, Farraj SA (2015) Biodiversity of marine scuticociliates (Protozoa, Ciliophora) from China: Description of seven morphotypes including a new species, Philaster sinensis spec nov. Eur J Protistol 51(2): 142-157.
- Xiong J, Wang G, Cheng J, Tian M, Pan X, et al. (2015) Genome of the facultative scuticociliatosis pathogen Pseudocohnilembus persalinus provides insight into its virulence through horizontal gene transfer. Scientific Reports 5(1): 15470.
- Whang I, Kang HS, Lee J (2013) Identification of scuticociliates (Pseudocohnilembus persalinus, longisetus, Uronema marinum and Miamiensis avidus) based on the cox1 sequence. Parasitol Int 62(1): 7-13.
- Huang YX, Wang S, Gao YQ, Chen JH, Wang XL, et al. (2021) Comparison of mitochondrial genome and development of specific PCR primers for identifying two scuticociliates, Pseudocohnilembus persalinus and Uronema marinum. Parasit Vectors 14(1): 318.
- Song JY, Kitamura S, Oh MJ, Kang HS, Lee JH, et al. (2009) Pathogenicity of Miamiensis avidus (syn. Philasterides dicentrarchi), Pseudocohnilembus persalinus, Pseudocohnilembus hargisi and Uronema marinum (Ciliophora, Scuticociliatida). Dis Aquat Organ 83(2): 133-143.
- Jones SR, G. Prosperi GP, Lapatra SE (2010) First Isolation of Pseudocohnilembus persalinus (Ciliophora: Scuticociliatida) From Freshwater-Reared Rainbow Trout, Oncorhynchus mykiss. The Journal of Parasitology 96(5): 1014-1016.
- Wei W, Chen K, Miao W, Yang W, Xiong J (2018) Pseudocohnilembus persalinus genome database - the first genome database of facultative scuticociliatosis BMC Genomics 19(1): 676.
- Kim SM, Cho JB, Lee EH, Kwon SR, Kim SK, et al. (2004) Pseudocohnilembus persalinus (Ciliophora: Scuticociitida) is an additional species causing scuticociliatosis in olive flounder Paralichthys olivaceus. Dis Aquat Organ 62(3): 239-244.
- Liu Y, Nan B, Duan L, Cheng T, Bourland WA (2019) A simple and rapid cryopreservation technique for ciliates: A Long‐Term Storage Procedure Used for Marine Scuticociliates. Journal of Eukaryotic Microbiology 66(5): 836-848.
- Weijie M, Chongnv W, Xuming P, Weixin J, Yuhang W, et al. (2020) TiO2 nanoparticles and multi-walled carbon nanotubes monitoring and bioremediation potential using ciliates Pseudocohnilembus persalinus. Ecotoxicol Environ Saf 187: 109825.
- OIE (2019) Use of animals in research and education. World Organization for Animal Health Paris, France.
- Weller T, Dammin G (1945) The acid-ether centrifugation and the zinc sulfate flotation techniques as methods for the recovery of the eggs of schistosoma mansoni. American Journal of Tropical Medicine 25(4): 367-374.
- Menu E, Mary C, Toga I, Raoult D, Ranque S, et al. (2018) Evaluation of two DNA extraction methods for the PCR-based detection of eukaryotic enteric pathogens in fecal samples. BMC Research Notes 11(1): 206.
- Boutellis M, Aissi K, Harhoura R, Drali T, Kernif F, et al. (2021) Tazerouti, first molecular characterization of blastocystis subtypes from animals and animal-keepers stool in Algeria. Comp Immunol Microbiol Infect Dis 78: 101695.
- Poirier P, Wawrzyniak I, Albert A, El Alaoui, Delbac F, et al. (2011) Development and evaluation of a real-time PCR assay for detection and quantification of blastocystis parasites in human stool samples: prospective study of patients with hematological malignancies. Journal of Clinical Microbiology 49(3): 975-983.
- Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33(7): 1870-1874.
- Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25(11): 1451-1452.
- Zhan Z, Li J, Xu K (2019) Ciliate environmental diversity can be underestimated by the V4 region of SSU rDNA: Insights from Species Delimitation and Multilocus Phylogeny of Pseudokeronopsis (Protist, Ciliophora). Microorganisms 7(11): 493.
- Schoenle A, Nitsche F, Werner J, Arndt H (2017) Deep-sea ciliates: Recorded diversity and experimental studies on pressure tolerance, deep sea research part I: Oceanographic Research Papers 128: 55-66.
- Zhao F, Xu K, Zhang D (2013) Spatio-temporal variations in the molecular diversity of microeukaryotes in particular ciliates in soil of the yellow river delta, China. The Journal of Eukaryotic Microbiology 60(3): 282-290.
- Goux X, Calusinska M, Fossépré M, Benizri E, Delfosse P (2016) Start-up phase of an anaerobic full-scale farm reactor-Appearance of mesophilic anaerobic conditions and establishment of the methanogenic microbial community. Bioresource Technology 212: 217-226.
- Foissner W (1987) New and little-known hypotrichous and colpodid ciliates (Protozoa: Ciliophora) from soils and mosses. Zoological Contributions 31(2): 187-282.
- Santos CC, Motta SP, Martins NS, Moreira AD, Alam NN, et al. (2020) Cryptosporidium spp. in Columba livia Gmelin, 1789 (Columbiformes: Columbidae) free-living pigeons from urban areas in Pelotas, Rio Grande do Sul, Brazil. J Parasit Dis 44(4): 877-881.
- Ramesh S, Soundararajan C, Subapriya S, Sokkalingam R, Muthukrishnan S (2018) Concomitant infection of Coccidiosis and Capillariasis in Fancy Pigeons (Columba livia)-A Case Report. Int J Curr Microbiol App Sc 7(12): 3701-3704.
- Peters A, Das S, Raidal SR (2020) Diverse trichomonas lineages in australasian pigeons and doves support a columbid origin for the genus trichomonas. Mol Phylogenet Evol 143: 106674.
- Collántes F, Fort MC, Ortega LM, Schares G (2018) Trichomonas, in parasitic protozoa of farm animals and pets. Springer 313-388.
- Toepfer EW (1964) Colpoda steinii in oral swabbings from mourning doves (Zenaidura macroura L). The Journal of Parasitology 50: 703.
© 2024, Amina Boutellis. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)













.bmp)
.jpg)
.png)
.jpg)














.png)

.png)



.png)






