- Submissions

Full Text
Cohesive Journal of Microbiology & Infectious Disease
Isolation and Characterization of Bacteriophages from Wastewater Sources on Enterococcus spp. Isolated from Clinical Samples
Yara Elahi1, Jamileh Nowroozi2 and Ramin Mazaheri Nezhad Fard3*
1Department of Genetics, Iran
2Department of Microbiology, Iran
3Department of Pathobiology, Iran
*Corresponding author: Ramin Mazaheri Nezhad Fard, Department of Pathobiology, Iran
Submission: March 29, 2021; Published: May 5, 2021

ISSN 2594-0190 Volume5 issues2
Abstract
In recent decades, enterococcal resistance to chemical antimicrobials has greatly increased. Furthermore, these chemicals include several side effects on the patients. Alternatively, researchers have investigated for novel treatments such as bacteriophages. Since no reports are available on effects of the bacteriophages on eukaryotic cells, these viruses can be good alternative solutions for multiple-resistant bacterial problem. Therefore, the major aim of this study was to investigate bacteriophages from municipal wastewaters affecting antimicrobial-resistant enterococci. The major aims of this study were to isolate and identify bacteriophages of on antibiotic-resistant clinical enterococci. Results indicated that bacteriophages could easily be isolated from wastewater sources. The three isolated bacteriophages were effective on clinical Enterococcus faecium as well as Streptococcus dysgalactiae ATCC 27957. Furthermore, bacteriophages were phenotyping studied. In general, bacteriophages belonged to Myoviridae, Siphoviridae and Inoviridae families of Caudovirales order. In conclusion, bacteriophages widely invade bacterial hosts and hence can be suggested as viable alternatives to current antimicrobials. Furthermore, (there is more in vivo studies needed to) to verify maximum effectiveness of the enterococcal bacteriophages.
Keywords: Enterococcus spp; Bacteriophages; Clinical samples; Wastewaters
Introduction
Enterococcus spp. are Gram-positive bacteria and gastrointestinal tract colonizers that opportunistically colonize wounds and bloodstream, causing life-threatening infections such as bacteremia and endocarditis [1,2]. These bacteria are particularly associated with Central Line Associated Bloodstream Infection (CLABSI), a type of Hospital Acquired Infection (HAI) that arises from use of central venous catheters. Enterococci are linked to 18% of CLABSIs in the United States [3]. The bacteria are part of the normal intestinal flora of mammals, birds and humans. Of all enterococcal species, Enterococcus faecalis and E. faecium are the most commonly identified species in human samples, whereas E. gallinarum and E. casseliflavus are less identified [4]. Enterococci are the cause of nosocomial infections, most frequently associated with intra-abdominal, pelvic, catheter, surgical, Central Nervous System (CNS) and Urinary Tract (UT) infections and endocarditis [5]. Enterococci are shown to include relatively high antibiotic resistances. It has been reported that enterococci from livestock and companion animals can be transmitted to humans via direct contacts [6]. In terms of public and animal health, it is important to prevent transmission of Multidrug-Resistant (MDR) enterococcal strains between animals or from animals to humans [7]. A previous study reported that animals treated with antibiotics in Intensive Care Unit (ICU) were sources or zoonotic transmitters of MDR Enterococcus species [8]. In USA, 35-75% of enterococcal infections are caused by E. faecium; from which, a majority are vancomycin-resistant [9]. In addition to threats posed by VRE, MDR enterococci serve as reservoirs for Horizontal Gene Transfer (HGT) of antibiotic resistance to other pathogens as verified by reports of van A transfer from Enterococcus species to Staphylococcus aureus [10].
Bacteriophages (phages) are viruses capable of infecting and replicating within the bacterial cells. They are the most abundant and ubiquitous entities on Earth, playing important roles in microbial physiology, evolution, population dynamics and therapeutics. Bacteriophages replicate through two primary life cycles or dynamic mechanisms, which are important for their therapeutic uses. Virulent or obligate lytic bacteriophages infect and quickly kill their bacterial hosts, whereas temperate or lysogenic bacteriophages may either stably integrate into their host genome or enter a lytic life cycle [11]. Phage therapy is described as direct administrations of lytic bacteriophages to patients to lyse bacterial pathogens causing clinical infections [12]. Recently, phage therapy has been interested as an alternative antimicrobial strategy to treat antibiotic-resistant biofilm-forming pathogens. For example, topical phage therapy is now considered as a good option in infections of burn wounds primarily caused by Pseudomonas aeruginosa and Staphylococcus aureus [13]. Therefore, the major aims of this study were to isolate and identify bacteriophages from municipal wastewaters on antibiotic-resistant clinical Enterococcus strains.
Materials and Methods
The following stages were carried out in this study to isolate and identify bacteriophages.
a) Isolation of the bacterial strains.
b) Sanger sequencing of the bacterial genome.
c) Bacterial antimicrobial susceptibility assessment.
d) Detection of bacteriophages.
e) Plaque purification and propagation.
f) Bacteriophage titration.
g) Bacterial genome extraction.
h) Bacteriophage genome typing.
i) Host specificity assessment of the bacteriophages.
j) Transmission electron microscopy.
Bacterial Strains
Bacterial strains were isolated from clinical patients referred to Imam Khomeini Hospital, Tehran, Iran, 2019-2020. The current study was approved by the Ethical Committee of Tehran University of Medical Sciences, Tehran, Iran (approval no. IR.TUMS.SPH. REC.1397.139). Relative information of the patients was collected using questionnaires. Blood samples were immediately cultured on bile esculin agar and incubated at 37 °C for 24h. Isolates were verified using morphological, biochemical and molecular techniques. First, isolates were Gram stained and studied using direct light microscope. Then, isolates were biochemically identified using arabinose fermentation test, salt tolerance test, optochin susceptibility test, CAMP test and PYR test. Molecular identification of the isolates was carried out through amplification of the tuf (elongation factor Tu) gene using Polymerase Chain Reaction (PCR) technique. An amplicon of each isolate was sequenced using Sanger sequencing method.
Sanger sequencing of the bacterial genome
To sequence the bacterial genome, a colony of the bacteria was dissolved in sterile distilled water using sterile microtube. The microtube was incubated at 90 °C for 30min to extract the genome. After centrifugation of the solution, Nano concentration of the extracted DNA was measured and the 260/280 and 260/230nm ratios were calculated using Drop One (Thermo Fisher Scientific, USA). Furthermore, genome was evaluated using agarose gel electrophoresis. The bacterial genome was sent for Sanger sequencing and the tuf gene partial sequence results were annotated in DNA Data Bank of Japan (DDBJ) under the accession numbers of LC580430 and LC580431.
Bacterial antimicrobial susceptibility assessment
The antimicrobial susceptibility patterns of the isolates were assessed using Kirby-Bauer method and the following antimicrobials of ceftriaxone (30μg), cefoxitin (30μg), clindamycin (2μg), erythromycin (15μg), linezolid (30μg) and vancomycin (30μg).
Detection of bacteriophages
Briefly, a colony of the bacterial sample was inoculated into 5ml of TSB liquid media and incubated at 37 ℃ overnight. Then, various wastewater samples were collected for bacteriophage isolation. Wastewater samples were centrifuged at 700g for 10min. After centrifugation, supernatant was filtered using 0.45-ml syringe filters. The bacterial samples in TSB were mixed with filtered sewage samples and 5ml of BHI broth media and incubated at 37 °C overnight. A colony of the bacterial sample was inoculated into TSB media and incubated at 37 °C overnight. Incubated culture was centrifuged at 700g for 10min. Supernatant was filtered through 0.45-ml syringe filters; then, 300μl of chloroform were added to the filtrate with agitation and set for 10-15min. This was centrifuged at 1600g for 5min and the supernatant was filtered through 0.45ml syringe filters. Then, 4.5ml of 0.75% TSA, 360μl of the filtered wastewater sample and 160μl of overnight bacteria were poured into a sterile microtube and set for 10min with agitation. This was mixed well with top agar and poured onto the plates. After 10min of setting at room temperature, plates were incubated at 37 °C overnight. After overnight incubation, plates were studied for the presence of the bacteriophage plaques.
Plaque purification and propagation
To purify bacteriophages, a plaque was selected from the plate and transferred into a SM buffer containing microtube using Pasteur pipettes. The microtube was centrifuged to precipitate debris and the liquid was filtered using syringe filters. The liquid was added to the broth media and after 24h, 360μl of TSB media and 160μl of the bacteria in logarithmic phase were added to 0.75% TSA top agar. After 24h, the previous steps were repeated three to five times to purify bacteriophages. Plaques achieved from the final purification steps were stored at 4 °C after centrifugation and filtration in SM buffer.
Bacteriophage titration
To prepare bacteriophage samples for TEM imaging and whole genome sequencing, bacteriophages were first diluted. For the titration of bacteriophages, nine microtubes were prepared for 100 to 10-8 serial dilutions. First, 900μl of SM buffer were added to each microtube. 100μl from the bacteriophage samples were transferred into the microtube of 100 dilution. Then, 100μl from this solution were transferred into the microtube of 10-1 dilution. This method was repeated to prepare all dilutions to 10-8. Then, 100μl of the diluted solution and 200μl of the overnight cultured bacteria were added to TSA top agar and poured onto the plates. After 24h, number of plaques in each plate was counted and the bacteriophage titers were calculated using the following formula:
C (pfu/ml) =n/(d × v (ml))
Where, C was the bacteriophage titer, n was the plaque count, d was dilution factor and v were the volume of dilution added.
Bacteriophage genome extraction
Briefly, 10μl of the bacteriophage solution were filtered and mixed with DNase 1 and RNase A. This was incubated at 37 °C for 30min and then mixed with 4μl of 20% PEG 6000. The mixture was incubated on ice for 1h and centrifuged for 30min. The supernatant was discarded, and the precipitate was dissolved in 600μl of SM buffer. Then, 25μl of phenol, 24μl of chloroform and 1μl of isoamyl alcohol were added to the solution. The solution was centrifuged for 20min and then mixed with 600μl of chloroform and recentrifuged for 20min. Then, 600μl of isopropyl alcohol were added to the mixture and incubated at -80 °C for 12min. After 20min of centrifugation of the solution and discarding of the supernatant, 700μl of ethyl alcohol were added to the precipitate. After 20min of centrifugation of the solution and discarding of the supernatant, the precipitate was dried under the hood for 10min and then dissolved in 50μl of TE buffer.
Bacteriophage genome typing
First, purity of the extracted bacteriophage genome was measured at 260/280 nm and 260/230 nm. Then, 1U of each DNase 1, RNase A and endonuclease S1 enzymes was added to 1μg of the extracted genomes and incubated at 37 °C for 2-3 h. In the next step, 10μl of each reaction were electrophoresed on 1% agarose gels and the resulting bands were analyzed under UV light.
Host specificity of bacteriophages
To investigate host specificity of the bacteriophages, the isolated bacteriophages were examined on Escherichia coli (ATCC 35218), Bacillus subtilis (ATCC 6633), Staphylococcus aureus (ATCC 29213), Salmonella enterica (ATCC 13076), salmonella typhimurium (ATCC 14028) and Streptococcus dysgalactiae (ATCC 27957). Standard strains. These strains were kindly provided by the Faculty of Veterinary Medicine, University of Tehran, previously characterized and used in other studies.
Transmission electron microscopy
To prepare bacteriophage samples for the electron microscopy, bacteriophage plaques were diluted to 10-8 in sterile microtubes using SM buffer. Microtubes were centrifuged and the supernatants were transferred to fresh sterile microtubes. Then, 200μl of each sample were used for bacteriophage staining. For the bacteriophage staining, 2% uranyl acetate dye and carbon-coated grids were used. After staining, samples were studied using Transmission Electron Microscopy (TEM) (Philips EM208S, Netherland) at 100 KV.
Result
In general, 25 Enterococcus strains were isolated from the clinical samples in this study. The enterococcal species, including E. faecalis, E. faecium and E. gallinarum, were isolated from blood samples. In this study, most of the isolated strains were resistant to vancomycin, erythromycin and clindamycin. Enterococcus faecium EntfacYE was sensitive to linezolid and resistant to vancomycin, erythromycin, clindamycin, ceftriaxone and cefoxitine. The tuf gene partial sequencing results verified primary characterizations of the isolated bacteria. Overall, three bacteriophages were isolated on one of the enterococcal isolates using two various wastewater sources of public ponds in Tehran (Figure 1). Two bacteriophages included isometric shapes, one with a long flexible tail (Siphoviridae) (Entfac.YE1) and another one with a non-flexible tail (Myoviridae) (Entfac.YE2) (Figures 2 & 3). The third bacteriophage was filamentous (Inoviridae) (Entfac.YE3) (Figure 4). The two tailed bacteriophages contained Double-Strand DNA (dsDNA) and the filamentous bacteriophage Single-Strand DNA (ssDNA) genomes. In host specificity assay, all the three bacteriophages were able to lysis Streptococcus spp. as well (Table 1).
Table 1: Characteristics of the isolated enterococcal bacteriophages.
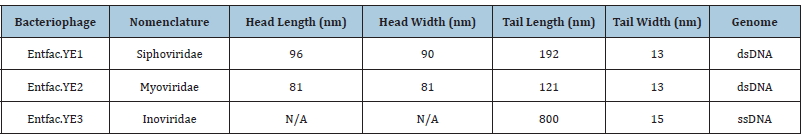
N/A: Not Applicable; dsDNA: Double-Stranded DNA; ssDNA: Single-Stranded DNA
Figure 1: Lysis of the Enterococcus species by bacteriophages.
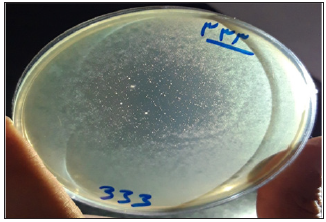
Figure 2: LEntfac.YE1 enterococcal bacteriophage an isometric head with flexible tail.

Figure 3:Entfac.YE2 enterococcal bacteriophage an isometric head with non-flexible tail.

Figure 4:Entfac.YE3 enterococcal bacteriophage a long filamentous bacteriophage.

Discussion
In this study, bacterial strains were isolated from human blood clinical samples. Of 25 isolated enterococci from blood samples, 14 E. faecium, 10 E. faecalis and one E. gallinarum were identified. In 2005, Mohanty et al. [14] identified 24 E. faecium, 7 E. feacalis and one E. gallinarum from 38 enterococci strains [14]. In 2019, Karna et al. [15] characterized four E. faecium and one E. faecalis in five enterococci isolates [15]. In another study, Rahbar et al. [16] isolated Enterococcus spp. from various wards of a university hospital in Tehran, Iran [16]. They identified 74 E. faecalis and 46 E. faecium from a total number of 120 enterococci isolates. In the current study, genetic identification of the enterococcal isolates was carried out using a pair of primers for enterococcal tuf gene and Sanger partial sequencing method. Previously, Li et al. [17] used the tuf gene as an appropriate target for the identification of Gram-positive cocci [17]. Furthermore, E. faecium is a gut commensal of humans and animals but also enlisted in global priority list of multidrug-resistant pathogens by the World Health Organization (WHO) [18]. In the current study, isolated E. faecium strains were resistant to vancomycin, erythromycin, clindamycin, ceftriaxone and cefoxitine. Based on the published studies, 70% of the isolated E. faecium strains from hospitals in Tehran were resistant to vancomycin and erythromycin [19]. In a similar study on clinical samples by Rahbar et al. [16] in Tehran, antibiotic resistance of enterococcal isolates to erythromycin was reported as 79% and to vancomycin as 51% [16]. In the present study, three various types of bacteriophages were isolated from a clinical strain of E. faecium using various wastewater sources. In previous studies, bacteriophages have been isolated from various wastewater sources such as raw, hospital and municipal wastewaters [20-22]. In the present study, all the isolated bacteriophages included lytic bacteriophages due to the clear plaques on agar plates [23]. Overall, three various bacteriophages were isolated on E. faecium, namely as EntfacYE1–3. These bacteriophages respectively belonged to Siphoviridae, Myoviridae and Inoviridae families of the Caudovirales order. Based on the literatures, these bacteriophages are mostly isolated on E. faecalis and E. faecium [24]. In a study in 2020, a Siphoviridae bacteriophage was isolated on E. faecalis, which was previously identified in 2012 [25,26]. In 2019 Wandro et al. [27] carried out a study on coevolution of E. faecium isolates from healthy human stools in absence and presence of Myoviridae bacteriophages [27]. In the current study, specificity assessment of the isolated bacteriophages demonstrated that the bacteriophages could lyse Streptococcus spp. as well as the Enterococcus spp. In fact, this broader host-specify of the isolated bacteriophages can be important due to the horizontal transfer of antimicrobial resistance genes and other virulence genes between various bacterial genera [28]. Moreover, this may be significant regarding treatment of a wider spectrum of the bacterial pathogens using a single bacteriophage, instead of a bacteriophage cocktail. Further characterization of the bacterial responses to bacteriophage infections is important for understanding how bacteriophages modulate bacterial physiology, which opens new horizons toward effective phage therapies against multiple-drug resistant bacteria [29]. Phage therapy, a promising alternative to antimicrobial treatment of bacterial diseases, is becoming further popular, especially due to the rapid spread of antimicrobial resistance in bacteria and recent restrictions on antimicrobial uses [16].
Conclusion
Bacteriophages are spread in the environment, especially in wastewaters. Nowadays, clinically isolated bacteria show multiple resistance to antibiotics, which have become serious problems for the medical specialists. Alternatively, bacteriophages can be used to solve these problems. Therefore, further clinical studies are necessary to investigate therapeutic properties of the bacteriophages on bacterial multiple-resistant strains. What is known about this subject? Several infectious diseases are caused by Enterococcus spp, especially by two common species of E. faecalis and E. faecium. Antibiotic resistance of these bacteria to common antibiotics, especially vancomycin, is increasing. Bacteriophages naturally infect enterococci. What this paper adds: Bacteriophages from wastewater sources are capable of lysing multiple-resistant enterococci from clinical samples. Most bacteriophages of the enterococcal isolates belong to head-tail bacteriophages. Identified bacteriophages are able to lyse S. dysgalactiae as well. Isolation of bacteriophages from wastewaters on clinical enterococci was considered in this research study. Since effects of bacteriophages on eukaryotic cells have not been reported so far and due to the extensive resistance of such isolates to common antimicrobials, bacteriophages are suggested as appropriate alternatives to routine antimicrobials for the treatment of enterococcal infections.
Acknowledgment
The authors thank staff within the Microbiology Laboratory for their kind helps. The current study was financially supported by a grant from the Deputy Dean for Research, Tehran University of Medical Sciences (grant no. 97-02-27-39571).
References
- Gilmore MS, Clewell DB, Ike Y, Shankar N (2014) Enterococci as indicators of environmental fecal contamination-in: enterococci: from commensals to leading causes of drug resistant infection. Massachusetts Eye and Ear Infirmary.
- Arias CA, Murray BE (2012) The rise of the Enterococcus: beyond vancomycin resistance. Nat Rev Microbiol 10(4): 266-278.
- Sievert DM, Ricks P, Edwards JR, Schneider A, Patel J, et al. (2013) Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the national healthcare safety network at the centers for disease control and prevention, 2009–2010. Infect Control Hosp Epidemiol 34(1): 1-4.
- Harwood VJ, Delahoya NC, Ulrich RM, Kramer MF, Whitlock JE (2004) Molecular confirmation of Enterococcus faecalis and faecium from clinical, faecal and environmental sources. Lett Appl Microbiol 38(6): 476-482.
- Domig KJ, Mayer HK, Kneifel W (2003) Methods used for the isolation, enumeration, characterization and identification of Enterococcus spp.: 2. Pheno and genotypic criteria. Int J Food Microbiol 88(2-3): 165-188.
- Bortolaia V, Guardabassi L (2015) Zoonotic transmission of antimicrobial resistant enterococci: a threat to public health or an overemphasised risk? Zoonoses-Infections Affecting Humans and Animals.
- Kataoka Y, Umino Y, Ochi H, Harada K, Sawada T (2014) Antimicrobial susceptibility of enterococcal species isolated from antibiotic-treated dogs and cats. J Vet Med Sci 76(10): 1399-1402.
- Ghosh A, Dowd SE, Zurek L (2011) Dogs leaving the ICU carry a very large multi-drug resistant enterococcal population with capacity for biofilm formation and horizontal gene transfer. PLoS One 6(7): e22451.
- Gilmore MS, Lebreton F, Van Schaik W (2013) Genomic transition of enterococci from gut commensals to leading causes of multidrug-resistant hospital infection in the antibiotic era. Curr Opin Microbiol 16(1): 10-16.
- Chang S, Sievert DM, Hageman JC, Boulton ML, Tenover FC (2003) Infection with vancomycin-resistant Staphylococcus aureus containing the vanA resistance gene. N Engl J Med 348(14): 1342-1347.
- Clokie MR, Millard AD, Letarov AV, Heaphy S (2011) Nature in phages. Bacteriophage 1(1): 31-45.
- Viertel TM, Ritter K, Horz HP (2014) Viruses versus bacteria novel approaches to bacteriophage therapy as a tool against multidrug-resistant pathogens. J Antimicrob Chemother 69(9): 2326-2336.
- Rose T, Verbeken G, De Vos D, Merabishvili M, Vaneechoutte M, et al. (2014) Experimental bacteriophage therapy of burn wound infection: difficult first steps. IJBT 4(2): 66.
- Mohanty S, Jose S, Singhal R, Sood S, Dhawan B, et al. (2005) Species prevalence and antimicrobial susceptibility of enterococci isolated in a tertiary care hospital of North India. Southeast Asian J Trop Med Public Health 36(4): 962.
- Karna A, Baral R, Khanal B (2019) Characterization of clinical isolates of enterococci with special reference to glycopeptide susceptibility at a tertiary care center of Eastern Nepal. Int J Food Microbiol Article ID: 7936156.
- Rahbar M, Majidpour A, Talebi M, Talebi TM (2016) Isolation and antibiotic susceptibility pattern among vancomycin resistant enterococci isolated from clinical samples of different parts of Rasoul-E-Akram Hospital. JAUMS 15(4): 404-413.
- Li X, Xing J, Li B, Wang P, Liu J (2012) Use of tuf as a target for sequence-based identification of Gram-positive cocci of the genus Enterococcus, Streptococcus, coagulase-negative Staphylococcus, and Lactococcus. Ann Clin Microbiol Antimicrob 11(1): 31.
- Chatterjee A, Willett JL, Nguyen UT, Monogue B, Palmer KL (2020) Parallel genomics uncover novel enterococcal-bacteriophage interactions. MBio 11(2): 3120-3139.
- Samadi H, Pirhajati MR, Pournajaf A, Omidi S, Moghimyan S, et al. (2015) An investigation of the vanA and vanB genes in Enterococcus faecalis and Enterococcus faecium strains isolated from the hospitalized patients in Shariati Hospital and evaluation of their antibiotic susceptibility. Qom Univ Med Sci J 9(3): 32-38.
- Imeni S, Akhavan SA, Soltan DM (2016) Isolating coli bacteriophage from raw sewage and determining its selectivity to the host cell. TB 15(1): 1-9.
- Torabi BP, Soltan DMM, Akbarzadeh S (2018) Isolation and specificity of Salmonella enteritidis bacteriophage from hospital sewage sample. RJMS 25(167): 1-9.
- Shokri D, Soleimani DA, Moayednia R, Mobasherizadeh S, Shirsalimian MS, et al. (2016) Isolation identification and evaluation of two lytic bacteriophages against clinical antibiotic-resistant strains of Pseudomonas aeruginosa from wastewater and hospital sewage of Isfahan City. SJIMU 23(7): 164-172.
- Budiarti S, Pratiwi RH, Rusmana I (2011) Infectivity of lytic bacteriophage to enteropathogenic Escherichia coli from diarrheal patients in Indonesia. J US-China Med Sci 8(5): 273-282.
- Arredondo AS, Top J, McNally A, Puranen S, Pesonen M, et al. (2020) Plasmids shaped the recent emergence of the major nosocomial pathogen Enterococcus faecium. MBio 11(1).
- Bhardwaj SB, Mehta M, Sood S, Sharma J (2020) Isolation of a novel phage and targeting biofilms of drug-resistant oral enterococci. J Infect Dis 12(1): 11-15.
- Horiuchi T, Sakka M, Hayashi A, Shimada T, Kimura T, et al. (2012) Complete genome sequence of bacteriophage BC-611 specifically infecting Enterococcus faecalis strain NP-10011. J Virol 86(17): 9538-9539.
- Wandro S, Oliver A, Gallagher T, Weihe C, England W, et al. (2019) Predictable molecular adaptation of coevolving Enterococcus faecium and lytic phage EfV12-phi1. Front Microbiol 9: 3192.
- Fard RM, Barton MD, Heuzenroeder MW (2010) Novel bacteriophages in Enterococcus spp. Curr Microbiol 60(6): 400-406.
- Kowalska JD, Kazimierczak J, Sowinska PM, Wojcik EA, Siwicki AK (2020) Growing trend of fighting infections in aquaculture environment-opportunities and challenges of bacteriophage therapy. Antibiotics 9(6): 301.
© 2021, Ramin Mazaheri Nezhad Fard. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)





























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)













.bmp)
.jpg)
.png)
.jpg)














.png)

.png)



.png)






