- Submissions

Full Text
Research in Medical & Engineering Sciences
5-Methylcytosine DNA Methylation Patterns among Gut Predominate Commensal Escherichia coli and Lactobacilli from the Balbas and Mazekh Domestic Sheep Breeds
Pepoyan AZ1,2*, Balayan MA1,2, Bezhanyan T2, Badalyan M1,2, Tsaturyan V2, Melnikov V3, Kamiya S4, Netrebov5 and Chikindas M6
1Armenian National Agrarian University, Armenia
2International Association for Human and Animals Health Improvement, Armenia
3Central Research Institute of Epidemiology, Russia;
4Kyorin University School of Medicine, Japan
5School of Environmental and Biological Sciences, Rutgers State University, USA
6Center for Digestive Health, NJ Institute for Food, Nutrition and Health, USA
*Corresponding author: Astghik Pepoyan, Food Safety and Biotechnology department, Armenian National Agrarian University, 74 Teryan, Yerevan 0009, Armenia
Submission: August 24, 2017; Published: eptember 08, 2017

ISSN : 2576-8816Volume1 Issue3
Abstract
The aim of current investigation was to compare the 5-methylcytosine level in genomic DNA of commensal Escherichia coli and lactobacilli from the widely bred in Armenia sheep breeds-Mazekh and Balbas. The results revealed statistically confirmed differences in5-methylcytosine methylation patterns of these bacteria in the bacterial isolates from Mazekh and Balbas sheep. The percentage of 5-methylcytosine of the genomic DNA was statistically higher for isolates from the Mazekh sheep in comparison with those of Balbas sheep (34.5±7.57 vs. 5.05±2.4, P <0.05). The Scatter Plot of relationship between cell surface hydrophobicity and genomic DNA’s 5-methylcytosine levels shows a moderately strong negative relationship between these characteristics of sheep gut isolates (Pearson R is -0.75). These results may be important for veterinary medicine and breeding programs.
Keywords:Commensals; Escherichia coli; Gut; Lactobacilli; Sheep
Introduction
At present, significant changes have occurred in the natural habitat of animals and humans. Ecologically unfavorable territories have appeared where a large number of people live and thousands of farm animals and birds are hold. Maintaining the quality of life in such areas is the most important task of veterinary medicine. Microbial biocenosis, and, especially, the biocenosis of the gastrointestinal tract is an important part in the well-being of animals and humans. It refers to the qualitative and quantitative composition of bacteria specific to each species. It is known that the formation and state of the gutmicrobiotais influenced by the breed characteristics of animals [1-3]. On the other hand it became apparent that the traditional view on bacterial population as clonal is not correct, and that phenotypic heterogeneity is common in bacteria. Divergent bacterial subpopulations are formed during adaptation to different environmental condition, including bacterial colonization of animals gut by commensals and pathogens. Epigenetic mechanisms can often be cause of such heterogeneity.
Epigenetic refers to biochemical modifications of chromatin that do not change the sequence but regulate gene expression and may be inherited. Several types of epigenetic modifications exist, such as DNA methylation, histone modification, and nucleosome positioning, and they can regulate a variety of processes, including transcription and protein binding to DNA. In most cases, DNA methylation is associated with gene silencing and is mediated by a family of enzymes called DNA methyltransferases [4,5]. The aim of current investigations was to compare the 5mC (5-methylcytosine) methylation levels of genomic DNA in commensal E. coli and lactobacilli from the widely bred in Armenia breeds of sheep – Mazekh and Balbas.
Materials and Methods
The study included 2-3 years old 15 Mazekh and 15 Balbas sheep from the Armenian farms. 30 E. coli and 30 lactobacilli isolates from the fecal microbiota of each sheep breed were investigated. Fecal materials were collected and analyzed as previously described [6]. Four predominant isolates found in the most diluted samples were grown and investigated. Bacteria were grown anaerobically according to Stepanyan and co-authors [7,8]. de-Man, Rogosa and Sharpe (MRS) Broth, Difco, (Fisher scientific) was used to propagate both Lactobacillus spp. and E. coli cells.
gDNA were extracted from bacterial cultures grown overnight using QIAamp® DNA Mini kit (Qiagen, Hilden, Germany) and the 5-mC methylation were identified by 5-mC DNA ELISA kit (Zymoresearch, California, USA) according to manufacturers’ protocols. The absorbance was measured at 450 nm on Stat Fax® 3300 (Awareness Technology, Inc, USA). The relationship between CSH and 5-mC methilation levels of isolates’ DNA was evaluated by the Pearson test (Excel 2010).
Results and Discussion
Gut infections and inflammation are frequently go together with dysbiosis. Inflammation induces a cascade of pro-inflammatory and anti-inflammatory molecules. The balance between these two groups of regulators controls cell death and repair of tissue damage. In recent years it has become apparent that gut bacteria produce many molecules which can counteract pro-inflammatory and anti-inflammatory pathways leading to activation or repression of immunity. Commensal bacteria can control the inflammatory process by altering the gut environment, changing the permeability barrier of the intestine, and by degradation of enteral antigens. Recent experiments have identified bacterial DNA, and unmethylated CpG motifs in particular, as another microbial stimulus which can be sensed by cells of the innate immune system, and induce the synthesis of an array of cytokines by immune cells [4,9]. DNA methylation plays a significant role in regulating gene expression, DNA replication and repair. Bacterial strains differ in DNA–methylation patterns. Approximately 35% of the bacterial DNAs contained N4-methylcytosine (4-mC), about 60% contained 5-mC, and about 90% had N6-methyladenine (6- mA) [10]. These modifications are performed for double-stranded DNA using methyltransferases or other methyl transfer machinery. Methylation of adenines may have broad roles in gene regulation and DNA replication in bacteria [11]. in comparison with eukaryotic cells: chromosome replication, nucleoid segregation, DNA repair, transposition of insertion elements [12], and transcription of specific genes is regulated by DNA adenine methylation (Dam), moreover, Dam methylation is required for virulence in pathogenic Enterobacteriaceae spp.>, including pathogenic E. Coli (Wion and Casadesús, 2006).
At the same time, cytosine can undergo modifications, forming 5-mC and its oxidized products 5-hydroxymethylcytosine (5-hmC), 5-formylcytosine (5-fC) and 5-carboxylcytosine (5-caC). 5-mC reduces and 5-hmC enhances DNA flexibility, and 5-caC does not have a measurable effect [13]. In bacteria, there are no known type I–III restriction-modification systems involving 5hmC or ghmC, though, several type IV systems that restrict DNA containing 5hmC and/ or ghmC have been described [14-16]. In E. Coli 6-mA and 5-mCbases are products of reactions catalyzed by three enzymes that, besides Dam, are specified by the host specificity (hsd) and DNA cytosine methylation (dcm) genes. DNA methylases isolated from E. Coli are site specific, they do not methylate DNA from E. coli in vitro, but they act on DNA from unrelated organisms.
Figure 1: Scatter plot of cell surface hydrophobicity - 5-methylcitosine relationship in sheep breeds of Mazekh and Balbas.
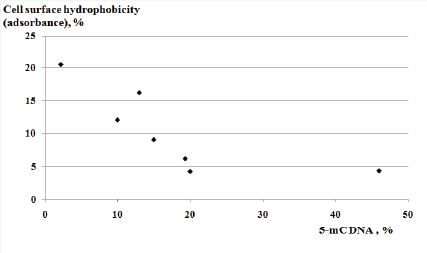
The results of our investigations revealed statistically confirmed differences between the 5-mC DNA methylation patterns of Mazekh and Balbas breeds’ bacterial isolates. The percentage of 5-mC DNA was statistically higher in investigated isolates from the Mazekh sheep in comparison with those of Balbas sheep (34.5±7.57 vs. 5.05±2.4, P<0.05). The Scatter Plot picture of relationship between cell surface hydrophobicity and genomic DNA’s 5-mC levels is presented in Figure 1. A moderately strong negative relationship between the 5-mC levels and CSH percentage were found for the seven randomly chosen isolates (Pearson R is -0.75) (Figure 1).
More research is needed to understand the role of these differences in the DNA methylation of gut commensals for animals’ immune status. Taking into account the above mentioned literature data, we hypothesized that the association of N6-methyladeninein eukaryotes and 5-mC- in bacteria might be important in hostbacteria interactions. At the same time these results may be important for the specific sheep husbandry/breeding programs.
References
- Lee WJ, Hase K (2014) Gut microbiota-generated metabolites in animal health and disease. Nat Chem Biol 10(6): 416-424.
- Gerritsen J, Smidt H, Rijkers GT, Vos DWM (2011) Intestinal microbiota in human health and disease: the impact of probiotics. Genes Nutr 6(3): 209-240.
- Zhang YJ, Li S, Gan RY, Zhou T, Xu DP, et al. (2015) Impacts of gut bacteria on human health and diseases. Int J Mol Sci 16(4): 7493-7519.
- Zhu FG, Marshall JS (2001) CpG-containing oligodeoxynucleotides induce tnf-alpha and il-6 production but not degranulation from murine bone marrow-derived mast cells. J Leukoc Biol 69(2): 253-262.
- Marinus MG, Casadesus J (2009) Roles of DNA adenine methylation in host-pathogen interactions: mismatch repair, transcriptional regulation, and more. FEMS Microbiol Rev 33(3): 488-503.
- Pepoyan AZ, Balayan MH, Manvelyan AM, Mamikonyan V, Isajanyan M, et al. (2017) Lactobacillus acidophilus INMIA 9602 Er-2 strain 317/402 probiotic regulates growth of commensal escherichia coli in gut microbiota of familial mediterranean fever disease subjects. Lett Appl Microbiol 64(4): 254-260.
- Stepanyan K, Balayan M, Vassilian A, Pepoyan A, Trchounian A, et al. (2007) Some Growth Peculiarities and Membrane characteristics of probiotic strains of Escherichia coli. Biochemistry (Moscow) Supplement Series A: Membrane and Cell Biology 1(4): 331-335.
- Balayan M, Mirzabekyan S, Isajanyan M, Pepoyan Z, Trchounian A, et al. (2010) Some peculiarities of growth and functional activity of escherichia coli strain from probiotic formula ASAP. International Journal of Biological and Medical Sciences 68: 670-675.
- Bogan JA, Helmstetter CE (1997) DNA sequestration and transcription in the oric Region of Escherichia coli . Mol Microbiol 26(5): 889-896.
- Ehrlich M, Wilson GG, Kuo KC, Gehrke CW (1987) N4-methylcytosine as a minor base in bacterial DNA. J Bacteriol 169(3): 939-943.
- Burgess DJ (2013) Epigenetics: Bacterial DNA Methylation Gets SMRT. Nat Rev Genet 14(1): 4-5.
- Wion D, Casadesús, J (2006) N6-methyl-adenine: an epigenetic signal for DNA-protein Interactions. Nat Rev Microbiol 4(3): 183-192.
- Ngo TT, Yoo J, Dai Q, Zhang Q, He C, et al. (2016) Effects of cytosine modifications on DNA Flexibility and Nucleosome Mechanical Stability. Nature Communications 7: 10813.
- Zheng Y, Karni CD, Xu D, Chin HG, Wilson G, et al. (2010) A Unique family of mrr-like modification-dependent restriction endonucleases. Nucleic Acids Res 38(16): 5527-5534.
- Bair CL, Black LW (2007) A Type IV modification dependent restriction nuclease that targets glucosylated Hydroxymethyl cytosine modified DNAs. J Mol Biol 366(3): 768-778.
- Xu SY, Corvaglia AR, Chan SH, Zheng Y, Linder P (2011) A Type IV modification-dependent restriction enzyme sauusi from staphylococcus aureus subsp. aureus USA300. Nucleic Acids Res 39(13): 5597-5610.
© 2017 Pepoyan AZ, et al. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)





























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)













.bmp)
.jpg)
.png)
.jpg)














.png)

.png)



.png)






