- Submissions

Full Text
Approaches in Poultry, Dairy & Veterinary Sciences
Serotyping of Salmonella Species in Poultry and Investigation of Antibiotic Susceptibilities
Kirkan Sukru*, Parin Ugur, Kutu Aysegul and Yuksel Hafize Tugba
Department of Microbiology, Adnan Menderes University, Turkey
*Corresponding author: Kirkan Sukru, Department of Microbiology, Faculty of Veterinary Medicine, Adnan Menderes University, Aydin, Turkey
Submission: May 22, 2018;Published: July 12, 2018

ISSN 2576-9162Volume4 Issue2
Abstract
In this study, 253samples were collected from chicken of Salmonella sp. suspected poultry farms in the western part of the Aegean Region from 01 September to 20 December 2016. After the necropsy process of the suspected chickens was carried out in the poultry diagnostic unit, the samples of the organs from the chickens were transferred in the cold chain to the laboratory. Inoculations were performed from organ samples to culture media. In conclusion, 43 isolates were detected as Salmonella sp, 4 (9.3%) isolates were found as S. Enteritidis and 39 (90.7%) isolates were found as S. Typhimurium. As a result of the antibiogram test, S. Enteritidis isolates were susceptible to Gentamicin and Ceftriaxon in the ratio of 100%, susceptible to Kanamycin and Tetracycline in the ratio of 50%, intermediate susceptible to Cefotaxime in the ratio of 75%, and resistant to Ampicillin and Penicillin in the ratio of 100%. S. Typhimurium (n=39) isolates were susceptible to Ceftriaxon in the ratio of 92%, Gentamicin in the ratio of 82%, Kanamycin and Cefotaxime in the ratio of 61%, Tetracycline in the ratio of 51% and resistant to Ampicillin in the ratio of 97%, Penicillin in the ratio of 100%.
Keywords: Poultry; Salmonella sp; Serotyping; PCR; Identification
Introduction
Salmonella infections in poultry, serotypes (especially S. Enteritidis and S. Typhimurium), which can be seen in different host hosts, with Pullorum Disease and Poultry Typhus caused by host-specific immobilized serotypes (S. entericasubsp. enterica) is caused by Salmonellosis and Paratyphoid infections. Paratyphoid factors have preventive measures in terms of public health because they can lead to significant zoonoses resulting in consumption of poultry products in humans. Bacteria of the Salmonella genus cause many acute and chronic disease-related infections in poultry, low yield and economic losses due to mortality [1].
Today, the detection of Salmonella serovars by standard laboratory methods is quite laborious and takes up to seven days, since it is based on selective culturing in selective media followed by biochemical and serological confirmation and identification of suspect colonies. Therefore, polymerase chain reaction (PCR) and PCR have provided significant advantages in terms of speed, sensitivity, specificity, reproducibility and other diagnostic methods based on this meta-analysis. Their use in identification of Salmonella in clinical specimens and food samples has increased. Another advantage of PCR is that the reaction is dependent on the use of a particular substrate or expression of certain antigens, and differences in the biochemical properties of strains with this feature prevent phenotypic variation and diagnostic errors that may result from lack of detectable antigens [2].
Antibiotics are still used despite the fact that they are protected against diseases through breeding studies in the rapidly developing poultry sector. Rapid development from the early twentieth century to the present day, when antibiotic treatment of infectious diseases began, has led to many antibiotics categorized according to their target effect structures in bacteria [3]. Transmission of resistance genes in resistant strains due to improper use of antibiotics can occur not only in pathogenic microorganisms, but also in endogenous microflora. Thus, during carcass contamination or ovulation, it can lead to colonization in human intestinal flora by passing through animal germs. It is inevitable that national action plans and legislation are continually updated according to country conditions, the establishment of standardized monitoring systems, the sharing of results with veterinarians/physicians, the training of intelligent antibiotics, and the prevention of the development of antibiotic resistance [4]. In this research, it was aimed to investigate the isolation of Salmonella enterica, fecal samples in chickens, biochemical and molecular methods of serotypes and sensitivity of isolates to various antibiotics.
Materials and Methods
Sample collection
A total of 253 samples from broiler were collected 01 September to 20 December 2016. After the necropsy process of the suspected chickens was carried out in the poultry diagnostic unit, the samples of the organs taken from the chickens were taken in the cold chain to the laboratory. As a result of the necropsy performed on the chicken samples taken from the poultry, Salmonella sp. isolation and identification were performed from the liver, spleen, heart and cecum organs. Animal Experiments Local Ethics Committee, dated 25.08.2016 and numbered 64583101/2016/141, declared that there was no ethical penalty.
Isolation and Identification
After necropsy of chicken; liver, heart, spleen and cecum organs were homogenized in 225ml of buffered peptone water and incubated at 37°C for 18-24 hours to provide pre-enrichment. On the second day after the pre-enrichment, 0.1ml of pre-enriched culture was inoculated into tube containing 10ml of Rappaport- Vassiliadis Soy broth (RVS and 0.1ml of the tube containing 10ml of Tetrathionate Buyyon (Muller-Kauffman), then incubated at 42°C for 18-24 hours of enrichment. In addition, the selective enrichment was completed by adding 0.1ml of the Selenit-F broth, which contained 10ml, to the 18-24 incubation at 37°C. At the end of the third day, a whole loop of liquid culture was taken inoculated onto XLT-4 Agar. After incubation at 37°C for 24-48 hours (ISO 6579/2002), Salmonella suspected colonies were inoculated onto Nutrient Agar and prepared for identification according to their biochemical characteristics [5].
API 20E Identification
For API 20E biochemical assays, isolates were inoculated into a 20-well plastic test tube. Five test tubes (ADH, LDC, ODC, H2S, URE) were closed with mineral oil for anechoic reactions while the three tubes (CIT, VP and GEL) were completely filled. After 24 hours incubation at 37°C, test strips were evaluated according to the reaction and colour changes indicated in (Table 1). The information obtained is processed in the api web database and the suspect columns are identified as a result of the program data.
Genotypic Identification
DNA extraction: DNA extraction for PCR in Salmonella strains isolated in our study was performed with Fermentas® DNA extraction kit.
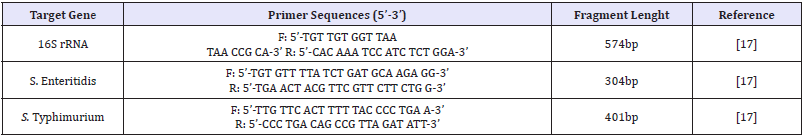
Primers: The primers used in the PCR method are shown in (Table 1)
Table 1: The primers used in the PCR method
PCR
16S rRNAPCR: In our study, genus-specific PCR procedures of isolating Salmonella isolates carrying the Salmonella enterica 16S rRNA gene were performed according to the protocol reported by Lin and Tsen [6]. PCR amplification for a sample in PCR reactions for the detection of 16S rRNA primer-specific products was performed in a final volume of 50μl with a final concentration of 1×Taq enzyme buffer solution, 50mM KCl, 1.5mM magnesium chloride (MgCl2), 200μmol each dNTP, 1μmol primer (for each), 0.5U Taq DNA polymerase (Genet Bio Exprime Taq DNA polymerase®) and 1μl 100ng template DNA. Thermal cycle and time diagram of PCR process used in 16S rRNA analyses was as follows initial denaturation at 94°C for 2min, 35 cycles consisting denaturation at 94°C for 20sec, annealing at at 54°C for 20 sec, extension at 72°C for 30 sec, and a cycle of final extension at 72°C for 4min.
Multiplex PCR for Salmonella Enteritidis/Salmonella Typhimurium: Specific PCR procedures for S. Enteritidis and S. Typhimurium serotypes were performed according to the protocol reported by Alvarez et al. [7]. PCR amplification for a sample in the PCR reaction was performed in a total volume of 25μl with a final concentration of 1×Taq enzyme buffer solution, 50mM KCl, 1.5mM magnesium chloride (MgCl2), 200μM each dNTP, 100nM primer (for each of the S. Enteritidis and S. Typhimurium specific primers), 1U Taq DNA polymerase (Genet Bio Exprime Taq DNA polymerase®) and 60pmol template DNA. Thermal cycle and time diagram was as follows: initial denaturation at 95°C for 2min, 30 cycles consisting denaturation at 54°C for 1min, annealing at 57°C for 1min, extension at 72°C for 2 min, and a cycle of final extension at 72°C for 5min.The 10μl amplified PCR products were detected by staining with 0.5μg/ml ethidium bromide after electrophoresis at 80 Volt for 40min in 2% agarose gel. The expected amplicon size was 574bp for Salmonella sp. (16S rRNA PCR), 304bp for S. Enteritidis, and 401bp for S. Typhimurium (Species-specific PCR).
Antibiogram
In our study, antibiotic susceptibility tests were performed with antimicrobial agents discs containing Penicillin G (P-10 U), Seftriaxon (CRO-30μg), Kanamycin (K-30μg), Cefotaxime (CTX-30μg), Ampicillin (AMP-10μg) Tetracycline(TE-30μg and Gentamicin (CN-10μg)(Oxoid®) used. The inhibition zone diameter around the antibiotic discs were measured and interpreted according to the standards of Clinical and Laboratory Standards Institute [8].
Results
Phenotypic findings
Clinical specimens were collected from liver, spleen, cecum and heart from chicken necropsy suspected of Salmonella infection. The inoculations were made on culture media for Salmonella Typhimurium-Salmonella Enteritidis isolation. Gram staining was performed on the colonies that grew upon incubation and Gram-negative colonies were passaged for identification tests. Biochemical tests on bacterial isolates were carried out by API 20E identification test. Salmonella sp. was isolated in 43 (17.0%) of 253 specimens. When the API 20E profiles of these isolates are examined, it is seen that all of them are identified as Salmonella sp. The API 20E test showed that 42 (98%) of the isolates was found as 6704552 profile,1 of Salmonella isolate were found as and 6704573 (2%) profile.
Figure 1: Genotype-specific PCR image carrying Salmonella enterica 16S rRNA gene.
M:100bp DNA ladder, 1: Negative control (Staphylococcus aureus ATCC 25923), 2-14: Salmonella enterica positive samples
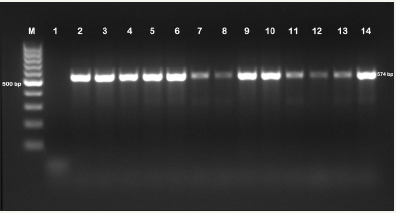
Figure 2: PCR images of S. Enteritidis and S. Typhimurium positive specimens.
M:100bp DNA ladder, 1: Negative control (Staphylococcus aureus ATCC 25923), 2-4-6-7-8-10-12: S. Typhimuriumpositive samples, 13: S. Enteritidispositive sample, 3-5-9-11: Salmonella negative samples.
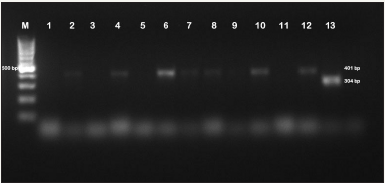
As a result of PCR in our study, all isolates (n=43) were identified as S. Enteric (Figure 1). Species-specific PCR for S. enterica isolates showed that 4 (9.3%) isolates were S. Enteritidis and 39 (90.7%) isolates were S. Typhimurium. Two (50%) of S. Enteritidis isolates were found to be broiler and the other 2 (50%) isolates belong to laying hens. Of the S. Typhimurium isolates, 31 (79.5%) belonged to the broiler and the other 8 (20.5%) belonged to the laying hens (Figure 2). In our study, 43 (17.0%) isolated Salmonella sp. were identified as S. enterica from clinical specimens. Of these isolates, 4 (9.3%) isolates were detected as S. Enteritidis and 39 (90.7%) isolates were detected as were found to be S. Typhimurium in conclusion of the genotypic identification.
Antibiogram
Table 2: The results of antibiograms of the isolates.
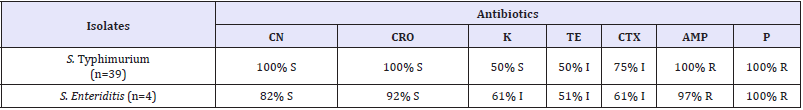
S: Susceptible, I: Intermediate susceptible, R: Resistant, CN: Gentamicin, CRO: Ceftiraxon, K: Kanamycin, TE: Tetracycline, CTX: Cefotaxime, AMP: Ampicillin, P: Penicillin
The results of antibiograms of the isolates identified in our study are shown in S. Enteritidis (n=4) isolates were susceptible to Gentamicin, Ceftriaxone (100%), Kanamycin and Tetracycline (50%); intermediate susceptible to Cefotaxime (75%) and resistant to Ampicillin and Penicillin (100%). S. Typhimurium isolates (n=39) were susceptible to Ceftriaxon (92%), Gentamicin (82%), Kanamycin and Cefotaxime (61%), Tetracycline 51%; resistant to Ampicillin (97%) and Penicillin (100%) (Table 2).
Discussion
In recent years, the prevalence of zoonotic gastrointestinal diseases worldwide has been noteworthy [9]. The incidence of reported Salmonella infections in humans in 2009 in European Union countries was reported to be 108,614 cases. The European Union reports that 23.7 percent of every 100.000 people have Salmonella cases. It has been found that 56% of the cases reported in 2009 are in the Czech Republic, Germany, England and Poland [10]. With the increased consumption of poultry meat, there is an increase in poultry-derived zoonotic diseases all over the world. Therefore, poultry meat contaminated with Salmonella and various products made from this group (sausage, salami etc.) are rarely cooked or uncooked eggs or products that are used in eggs are very dangerous for public health [11,12]. Scaallan et al. [13] found that 5.5 million (59%) of the 9.4 million foodborne infections in the United States were viral agents; 3.6 million (39%) were bacterial agents and 0.2 million (2%) were parasitic agents. It has been reported that approximately 1 million (11%) of the infections are caused by nontyphoid Salmonella sp., 42000 are supported by laboratory results, 19000 cases are hospitalized and 400 deaths have been reported.
The prevalence of Salmonella serovars in poultry shows differences between countries and years. Some serovars are important for countries over a period of time, sometimes disappearing without any symptoms. In history, S. Enteric serovar Typhimurium is the serovar with the highest prevalence isolated from the poultry. In England between 1968 and 1973, S. Typhimurium constituted 40% and S. Enteritidis constituted 6% of infections [14]. In the United States in 1990, 23.431 faecal specimens were collected from 406 laying farms and isolation of S. Enteritidis was 24% [15].
The diagnosis of Salmonella infections in poultry is traditionally carried out using selective media and characterization of suspected colonies by biochemical and serological tests. However, the standard laboratory procedures performed with these methods (cultureidentification) are completed in a very long period of time such as 4-8 days. In addition, Salmonella isolation cannot be performed in carrier animals and in some cases (presence of microorganisms inhibiting Salmonella in clinical specimens, antibiotic use etc.) in clinical specimens is small in number. For these reasons, much faster and more sensitive methods were needed, and more selective culture methods, DNA hybridization tests and immunoglobulin test were developed for this purpose. PCR can amplify the target DNA segments that can detect Salmonella present in clinical and environmental samples with a high sensitivity and specificity [16]. In their study, Alverez et al. [7] reported that in Spain, multiplex PCR was developed to detect five most important Salmonella serotypes and phage types simultaneously. Mir et al. [17] investigated the outbreaks and serotype diversity of Salmonella isolates in various poultry species using the 16 S rRNA genus-specific PCR technique in their study in India and reported that they used them to detect common serovars such as S. Enteritidis, S. Typhimurium. Çarlı et al. [18] reported that, after selective enrichment, they were able to rapidly and reliably detect Salmonella species by PCR, both experimentally and clinically. Türkyılmaz et al. [19] found Salmonella sp. as 29 (6.3%) and S. Enteritidis as 19 (4.1%) in 460 cloacal swab samples and reported S. Enteritidis in Salmonella sp. isolates as 65.5% in Aydin province of Turkey.
Various studies have been conducted worldwide on the isolation and serotyping of Salmonella species. Among the studies conducted, different prevalence rates of Salmonella infections and different numbers of Salmonella serovars have been reported in various poultry species (chicken, duck, turkey etc) in India [20]. Mir et al. [17] in their study of the Salmonella isolates in various poultry species in India to investigate the outbreak and serotype diversity; 327 (6.3%) Salmonella enterica isolates were isolated from 507 chickens, ostriches and ducks in total, 202 of which were identified from sera and 305 were fecal; 9 were S. Enteritidis, 5 were S. Typhimurium, 4 were S. Virchow. Dookeran et al [21], in their study of broiler poultry; They examined 64 poultry houses and found that 50% of these poultry houses were Salmonella sp. positive and reported a prevalence rate of 6-11% in the poultry houses.
In another Australian study, Lassnig et al. [22] tested 363 strains, each of which contained at least 5000 broilers, 28 of which (7.7%) were identified as Salmonella sp. and S. Enteritidis (1.7%), S. Typhimurium (0.6%), S. Montevideo (4.1%) and S. infantis (0.6%). In a study of Salmonella species in chicken faeces, internal organs and carcasses investigated by both PCR and standard culture method, Oliveira et al. [23] examined 64 drag swab samples against Salmonella and reported that they isolate Salmonella from 16 drag swab samples (25%). As a result of the genotypic identification of 43 (17.0%) isolates of Salmonella sp. isolated in our study, it was found that they were S. enterica and that 4 (9.3%) of these isolates were S. Enteritidis and 39 (90.7%) of these isolates were S. Typhimurium. It is seen that the ratio of S. Typhimurium is locally high in line with the results we have obtained.Antibiotics are widely used in the treatment of infections caused by Salmonella in poultry (typhoid, paratyphoid), resulting in resistance to antibiotics in Salmonella strains in intestine and other tissues of clinical or latent infected animals. Antimicrobial resistance is an undesirable side effect. The use of antimicrobials leads to the selection of clones of resistant bacteria (pathogenic, commensal or environmental bacteria) in humans and animals; threatens human health by altering the structure of the microbial population and accelerating the evolutionary process [24].
Antibiotics applied unconsciously, used for therapeutic and prophylactic purposes, can also be used to encourage growth in broiler production. Some antibacterial, such as chloramphenicol, tetracycline, ampicillin, enrofloxacin, neomycin, which are used in antibacterial treatment, cause the Salmonella agents to become resistant by inhibiting the beneficial flora of the nervous system. These bacteria play a role in the transfer of resistance genes and cause severe infections affecting the food chain when spreading in nature [25]. In addition, in transferring the antibiotic resistance of human origin to humans; antibiotic-resistant microorganisms in animal products have been reported to reach human beings through foods that are consumed without being sufficiently heat-treated, to be used in vegetative production of animal fertilizers containing resistant microorganisms or by animal wastes, and to be consumed by humans [26]. Similar studies have been carried out by different researchers on antibiotics and antibiotic resistance used in poultry breeding in the world and in our country. According to the EFSA 2013 report, Salmonella resistance from broiler chickens has been reported in 13 European countries including Austria, Denmark, France, Germany, Greece, Hungary, Ireland, Italy, Slovakia, Portugal, the Netherlands and Spain and resistance rate was found as 12.5% against Ampicillin, Cefloxacin, Ciprofloxacin, Gentamicin, Nalidixic acid, Sulfonamide and Tetracycline.
The highest resistance rate in the study was reported against Tetracycline and Sulfonamide, respectively, with rates of 42.2% and 42%. In Europe, the Tetracycline resistance of Salmonella strains was 31% and Sulfonamide resistance was 38%. In accordance with the results obtained in our study, it was determined that all Salmonella isolates (n=43) were approximately 100% resistant to Ampicillin and Penicillin and 80-100% susceptible to Gentamicin and Ceftriaxone. Salmonella species isolated from poultry are remarkably resistant against antibiotics in different shapes and frequency. Here, it is indicated that the activity of the resistanceencoding, conjugative or transferable plasmids is one of the most important factors in resistance spreading. However, differences in resistance have been reported to vary depending on the species and type of poultry, country or region, years, Salmonella serovar from one farm to another, laying eggs or broilers, and the mechanism of action of antibiotics [4].
Conclusion
Salmonella serovars have the ability to colonize and disease in many animals, especially in poultry. Due to its vertically contagious nature, it can cause food poisoning in people who consume poorly cooked poultry meat and eggs. In order to prevent and control the spread of Salmonella, monitorization of the disease agent periodically with bacteriological, serological and molecular methods with biosecurity measures is essential. Proper identification of isolates and treatment should be appropriate. In addition, extensive Salmonella serotype distribution and antibiotic susceptibility differ from region to region, so broad range epidemiological studies should be useful for future aspects.
Acknowledgement
This research was supported by the Adnan Menderes University Scientific Research Projects Committee (code No. VTF-17009).
References
- Sareyyüpoğlu B (2010) Kanatlı Salmonella Enfeksiyonlarının Konvansiyonel ve Moleküler Yöntemlerle Teşhisi. Türkiye Klinikleri 1(2): 69-79.
- Soumet C, Ermel G, Rose V, Rose N, Drouin P, et al. (1999) Identification by a multiplex PCR-based assay of Salmonella Typhimurium and Salmonella Enteritidis strains from environmental swabs of poultry houses. Lett Appl Microbiol 29(1): 1-6.
- Kaya S (2013) Kemoterapötikler. Veteriner Farmakoloji Medisan Yayınları, Ankara 322-665.
- Filazi A, Dikmen B, Kuzukıran Ö (2015) Kanatlılarda Antibiyotik Direnci. Türkiye Klinikleri Veteriner Bilimleri Farmakoloji ve Toksikoloji Özel Sayısı 1 (2): 42-51.
- OIE Manual of Diagnostic Tests and Vaccines for Terrestrial Animals.
- Lin JS, Tsen HY (1999) Development and use of polymerase chain reaction for the detection of Salmonella Typhimurium in stool and food samples. J of Food Prot 62(10): 1103-1110.
- Alvarez J, Sota M, Vivanco BA, Perales I, Cisterna R, et al. (2004) Development of a Multiplex PCR technique for detection and epidemiological typing of Salmonella in human clinical samples. J Clin Microbiol 42(4): 1734-1738.
- CLSI National Committee for Clinical Laboratory Standarts (M11) (2017) Performance Standards for Antimicrobial Susceptibility Testing. 27th Informational Supplement, Wayne.
- Makaya PV, Matope G, Pfukenyi DM (2012) Distribution of Salmonella serovars and antimicrobial susceptibility of Salmonella Enteritidis from poultry in Zimbabwe. Avian Pathol 41(2): 221-226.
- European Food Safety Authority (EFSA) (2011) The European Union Summary Report on trends and sources of zoonoses, zoonotic agents and foodborne outbreaks in 2009. EFSA Journal 9: 2090-2477.
- Barrow PA (1991) Experimental infection of chickens with Salmonella enteritidis. Avian Pathol 20(1): 145-153.
- Bilgehan H(2004) Klinik Mikrobiyolojik Tanı. Fakülteler Kitabevi Barış Yayınları, İzmir 425-455.
- Scaallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA (2011) Foodborne illness acquired in the United States - Major Pathogens. Emerg Infect Dis 17(1): 7-15.
- Sojka WJ, Wray C, Brand TF (1997) Agglutinins to common Salmonellae in the sera of apparently healthy sheep. Br Vet J 133(6): 615-622.
- Poppe C, Irwin RJ, Forsberg CM, Clarke RC, Oggel J (1991) The prevalence of Salmonella Enteritidis and other Salmonella spp. among Canadian registered commercial layer flocks. Epidemiol Infect 106(2): 259-270.
- Van Asten AJ, Van Dijk JE (2005) Distribution of “classic” virulence factors among Salmonella spp. FEMS Immunol Med Microbiol 44(3): 251-259.
- Mir IA, Kashyap SK, Sunil M (2015) Isolation, serotype diversity and antibiogram of Salmonella enterica isolated from different species of poultry in India. Asian Pacific Journal of Tropical Biomedicine 5(7): 561- 567.
- Çarlı KT, Ünal CB, Caner V, Eyigör A (2001) Detection of chicken feces by a combination of Tetrathionate Broth Enrichment, Capillary PCR, and Capillary Gel electrophhoresis. J ClinMicrobiol 39(5): 1871-1876.
- Türkyılmaz S, Savaşan S, Kırkan Ş, Kaya O (2007) Tavuklarda Salmonella Enteritidis infeksiyonlarının bakteriyolojik ve serolojik yöntemlerle teshisi. İstanbul Üniversitesi Veteriner Fakültesi Dergisi 33: 23-34.
- Ramya P, Madhavarao T, Rao LV (2012) Study on the incidence of Salmonella Enteritidis in poultry and meat samples by cultural and PCR methods. Vet World 5(9): 541-545.
- Dookeran MM, Baccus-Taylor GSH, Akingbala JO, Tameru B, Lammerding AM (2012) Transmission of Salmonella on broiler chickens and carcasses from production to retail in Trinidad and Tobago. Journal of Applied Business Research 1: 78-84.
- Lassnig H, Much P, Schliessnig H, Osterreicher E, Kostenzer K, et al. (2012) Prevalence of Salmonella spp. in Austrian broiler flocks in the context of the EU-wide baseline survey 2005-2006. Berl Münch Tierärztl Wochenschr 125(3-4): 129-137.
- Oliveira SD, Santos LR, Schuch DM, Silva AB, Salle CT, et al. (2002) Detection and identification of Salmonellas from poultry related samples by PCR. Vet Microbiol 87(1); 25-35.
- Şahan Ö, Aral EM, Aden MMA, Aksoy A, Yılmaz Ö, et al. (2016) Türkiye’deki broyler tavuk işletmelerinden izole edilen Salmonella serovarlarının antimikrobiyel direnç durumu. Ankara Üniv Vet Fak Derg 63: 1-6.
- Mølbak K, Baggesen DL, Aarestrup FM, Ebbesen JM, Engberg J, et al. (1999) An outbreak of multidrug-resistant Salmonella enterica serotype Typhimurium DT104. N Engl J Med 341(19):1420-1425.
- European Food Safety Authority (EFSA) (2013) The European Union Summary Report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals, and food in 2011. EFSA Journal 11(5): 3196.
© 2018 Kırkan Sukru. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)





























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)













.bmp)
.jpg)
.png)
.jpg)














.png)

.png)



.png)






