- Submissions

Full Text
Advancements in Bioequivalence & Bioavailability
Bioinformatics Programming for Bioavailability Analysis of Sequence Patterns in Public Genomic Databases
Changsu Dong1, Roy Lee1, Joseph Sayad2, and Konstantinos Krampis1,2,3
1 Belfer Research Building, Weill Cornell Medical College and Hunter College, USA
2 Department of Biological Sciences, Hunter College, USA
3 Department of Physiology and Biophysics, Cornell University, USA
*Corresponding author: Konstantinos Krampis, Department of Physiology and Biophysics, Institute for Computational Biomedicine, Weill Cornell Medical College, Cornell University, New York, USA
Submission: April 19, 2018; Published: May 10, 2018

ISSN 2640-9275 Volume1 Issue2
Abstract
In this study we present a novel bioinformatics software for the analysis of bioavailability of short amino acid peptides, in various proteins found across four phylogenetic kingdoms of Archaea, Bacteria, Mammals and Plants. In order to assess bioavailability of these peptides, we have used a set of large-scale protein databases from the National Center for Biotechnology Information, and the Basic Local Alignment Search Tools (BLAST+) search program. In our results we present the counts of peptides matches across the phylogenetic kingdoms, and also in further detail for Gram positive or negative bacteria. Our bioinformatics software is written in Python and is made available within this publication as freely available for academic and non-profit use.
Results
In order to understand the prevalence of the short peptide patterns in various proteins across different kingdoms (Archaea, Bacteria, Mammals, and Plants), we performed a set of pattern searches using sequence alignments to the databases of the National Center for Biotechnology Information [1]. Specifically, we used the Basic Local Alignment Search Tools (BLAST+) with a local copy the NCBI databases, and searched the databases with queries of peptide sequence patterns through the BLASTP protein alignment subprogram. For our database searches, we used a set of peptides sequences each six amino acids long, which were first described in [2], in addition to a set of twelve amino acid peptides described in [3]. According to these studies, these small protein peptides are factors in a range of diseases ranging from Parkinson’s and Alzheimer’s, to diabetes, mainly due to their role in formation of oligomer aggregates leading to amyloid plaques. The innovation of our study was the use of a powerful computer server in our laboratory, in combination with a novel computer code written in Python (Methods Section), to data mine the complete set of peptide matches from the BLAST+ to the NCBI databases. These databases contain a complete, non-redundant collection of reference genome sequences representative of all major organisms and phylogenetic tree clades. Using the NCBI FTP Site, we downloaded the Archaea, Bacteria, Mammals, and Plants reference databases (Table 1) in FASTA format [4]. Each kingdom had multiple FASTA files for each genome included in the kingdom, which were concatenated into a single file for each kingdom. Following this, the single files were searched for identities to the peptides from the selected studies described above using the BLAST+ programs, which were run with the parameters described in the Methods section.
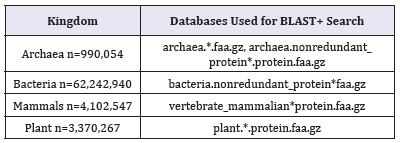
Table 1: Protein databases from NCBI used in our study.
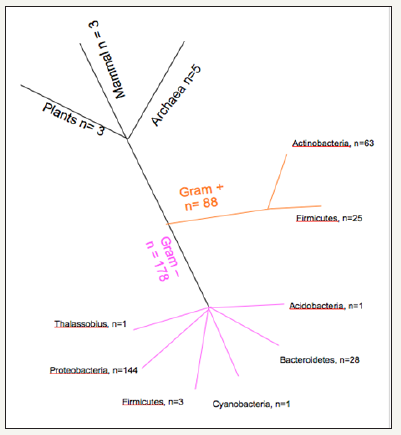
In our results from the BLAST+ searches of the different short peptides we observed 3 matches in the Plants and Mammals phylogenetic kingdoms, and 5 matches for the Archaea (Figure 1). The most numerous matches were for the Bacteria (266 matches), which was expected given the large number of species within this phylogenetic kingdom. In order to further clarify the results, we separated the Bacterial species in Gram+ (88 hits) or Gram- (178 hits). With this, more detailed numbers are presented for the different Phyla of the Bacteria in each Gram category. For clarity, the results were also visualized in tree format (Figure 1), demonstrating the number of matches across the different phylogenetic clades.
Figure 1: Number of matches by BLAST+ searches across the different phylogenetic kingdoms.
Methods
We performed four separate BLAST+ searches in the Archaea, Bacteria, Plants and Mammals phylogenetic kingdoms Table 1, using the BLASTP sub-program for peptide alignment to the database. Before performing BLASTP, we formatted the FASTA files (.faa) that were downloaded from NCBI, and produced files in the format required (.pal, .pni) for BLASTP database search. Towards this, we used the makeblastdb command, which takes a FASTA file as input and outputs a database ready for BLASTP. makeblastdb -in all_plantBlast.faa -parse_seqids –db type prot. The exact command parameters used were the ones shown below (with the Plant database as example, the .pal / .pni suffixes of the database files are not required in the command): blastp -outfmt 5 -query peptide. fa -word_size 2 -matrix=PAM30 -db all_plant Blast -out Plant.xml -evalue 100000000000 -max_target_seqs 500 -qcov_hsp_perc 100.
As seen in the command above, we set the BLASTP results to be written in the output file (“-out”) in extensible Markup Language (XML) format. The reason was that his format is standardized, making it easy to perform further analysis and data mining of the BLASTP results, using a programming language such as xml. etree in Python 2.7 shown on the “Code Insert” section below. The developed code reads the complete XML BLASTP output, in addition to filtering and counting matches for the database that contain any of the keywords related to the specific functionality of peptides (lines 16-20 of the code). Finally, the code prints the output, from which we counted the number of hits per kingdom presented in (Figure 1) in the results.
Bioinformatics Code
from xml.etree import Element Tree
#This program parses XML output from blast
#Input include 1) XML input
#Output 2) Output prints
# 2a) Blast Matches that are non-nucleotide binding
# 2b) Blast Matches that are nucleotide binding
def blast ExactMatch(file Name, hit Seq):
root = ElementTree.parse(fileName).getroot()
rootSub1 = root.getchildren()
iterations = rootSub1[8].getchildren()
blastOutputIterations = iterations[0].getchildren()
IterationHits = blastOutputIterations[4].getchildren()
Nucleotide Key Words = [‘SYNTHET’,’NUCLEOTID’,’ABC TRANSPORTER’,’PHOSPHO’,’ADP’,’AMP’,’ATP’,’ATP BINDING’,’ATPDEPENDENT’,’ ATPASE’,’CAMP’,’CDP’,’CGMP’,’CMP’,’COENZYME A’,’CTP’,’CTP
BINDING’,’CTP-DEPENDENT’,’DNA BINDING’,’DNA REPAIR’,’FAD’,’FADH 2’,’GDP’,’GMP’,’GTP’,’GTP
BINDING’,’GTP-DEPENDENT’,’GTPASE’,’HELICASE’,’NAD +’,’NADH’,’NADP +’,’NADPH’,’NUCLEOTIDE
BINDING’,’RNA BINDING’,’TRNA BINDING’,’UDP’,’UMP’,’UTP’,’UTP BINDING’,’UTP-DEPENDENT’]
TotalFullHits =0
TotalFullHitsNucleotide = 0
TotalAnyHits = 0
HitIDs = []
for hitDescr1 in IterationHits:
TotalAnyHits = TotalAnyHits ++ 1
hitDescr2 = hitDescr1.getchildren()[5]
hitDescr3 = hitDescr2.getchildren()[0]
Hsp_score = hitDescr3.findtext(‘Hsp_score’)
Hsp_qseq = hitDescr3.findtext(‘Hsp_qseq’)
Hsp_hseq = hitDescr3.findtext(‘Hsp_hseq’)
Hsp_midline = hitDescr3.findtext(‘Hsp_midline’)
Hsp_evalue = hitDescr3.findtext(‘Hsp_evalue’) Hit_num = hitDescr1.findtext(‘Hit_num’)
Hit_id = hitDescr1.findtext(‘Hit_id’)
Hit_def = (hitDescr1.findtext(‘Hit_def’)).upper()
NucleotideProtein = “NO “
if any(x in Hit_def for x in NucleotideKeyWords):
NucleotideProtein = “YES”
if Hsp_hseq==hitSeq and “HYPOTHETICAL PROTEIN”
not in Hit_def:
print “Hit Number:” + Hit_num
print “Hit ID:” + Hit_id
HitIDs.append(Hit_id)
print “Hit Def:” + Hit_def
print “Nucleotide Binding Protein:” + NucleotideProtein
print “Hit Accession:”+ hitDescr1.findtext(‘Hit_ accession’)
print “Hsp Midline:” + Hsp_midline
TotalFullHits = TotalFullHits + 1
print “Hsp_score:”+Hsp_score
print “Hsp_qseq:”+Hsp_qseq
print “Hsp_hseq:”+Hsp_hseq
print “Hsp_evalue:”+Hsp_evalue
if NucleotideProtein == “YES”:
TotalFullHitsNucleotide = TotalFullHitsNucleotide + 1
print “****************************************”
print “Total Blast Full Hits:”+str(TotalFullHits)
print “Blast Full Hits & Nucleotide:”+
str(TotalFullHitsNucleotide)
FullNonNuc = TotalFullHits - TotalFullHitsNucleotide
print “Blast Full Hits & Non-Nucleotide:”+ str(FullNonNuc)
def appendToFile(textFile,strToAppend):
with open(textFile,’a’) as file_object:
file_object.write(strToAppend)
def deleteLastLine(textFile):
lines = open(textFile).readlines()
open(textFile,’w’).writelines(lines[:-1])
def seqCount(fastaInput):
appendToFile(fastaInput,”>”)
with open(fastaInput) as file_object:
TotalNumberAA = 0
TotalNumberSeq = -1
for line in file_object:
lineFirstChar = line[0]
seqCount = 0
seq = ‘’
if line[0] == ‘>’:
try:
seqHeader = seqHeader2
except:
seqHeader = line
seqHeader2 = ‘’
print seqHeader
while (lineFirstChar!=’>’):
seq = seq + line
line = next(file_object)
lineFirstChar = line[0]
seqHeader2 = line
TotalNumberSeq = TotalNumberSeq + 1
seqHeader = seqHeader2
if seq!=’’:
print seq
ProteinLength = len(seq)-1
TotalNumberAA = TotalNumberAA + ProteinLength
print ProteinLength
print “************************************** *****”
print “TotalAAinDB:”+str(TotalNumberAA)
print “TotalSeqinDB:”+str(TotalNumberSeq)
deleteLastLine(fastaInput)
def createFastaFromRef(filterFAA,refDBFAA): header = “>”
with open(refDBFAA) as oldfile, open(filterFAA, ‘w’) as newfile:
for line in oldfile:
if any(HitID in line for HitID in HitIDs): #HitID are the elements from the forloop in this line
newfile.write(line)
myLine = next(oldfile)
while (myLine[:1]!=”>”):
newfile.write(myLine)
myLine = next(oldfile)
Acknowledgement
We thank all members of the Bioinformatics Core Infrastructures Lab for their useful feedback and suggestions during the preparation of the manuscript. This research was supported by the Center for Translational and Basic Research grant from National Institute on Minority Health and Health Disparities (G12 MD007599) and Weill Cornell Medical College-Clinical and Translational Science Center (2UL1TR000457-06).
References
- McGinnis S, Madden TL (2004) Blast: At the core of a powerful and diverse set of sequence analysis tools. Nucleic acids Research 32(2): 20- 25.
- Lee M, Wang T, Makhlynets OV, Wu Y, Polizzi NF, et al. (2017) Zincbinding structure of a catalytic amyloid from solid-state NMR. Proc Natl Acad Sci 114(24): 6191-6196.
- Bernstein SL, Dupuis NF, Lazo ND, Wyttenbach T, Condron MM, et al. (2009) Amyloid-β protein oligomerization and the importance of tetramers and dodecamers in the aetiology of Alzheimer’s disease. Nat Chem 1(4): 326-331.
- Lipman DJ, Pearson WR (1958) Rapid and sensitive protein similarity searches. Science 227(4693): 1435-1441
© 2018 Konstantinos Krampis. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)





























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)













.bmp)
.jpg)
.png)
.jpg)














.png)

.png)



.png)






